Identification of a novel Drosophila gene, beltless, using injectable embryonic and adult RNA interference (RNAi)
- PMID: 12914675
- PMCID: PMC194572
- DOI: 10.1186/1471-2164-4-33
Identification of a novel Drosophila gene, beltless, using injectable embryonic and adult RNA interference (RNAi)
Abstract
Background: RNA interference (RNAi) is a process triggered by a double-stranded RNA that leads to targeted down-regulation/silencing of gene expression and can be used for functional genomics; i.e. loss-of-function studies. Here we report on the use of RNAi in the identification of a developmentally important novel Drosophila (fruit fly) gene (corresponding to a putative gene CG5652/GM06434), that we named beltless based on an embryonic loss-of-function phenotype.
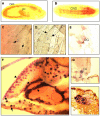
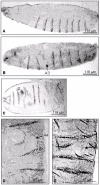
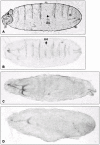
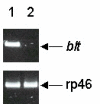
Results: Beltless mRNA is expressed in all developmental stages except in 0-6 h embryos. In situ RT-PCR localized beltless mRNA in the ventral cord and brain of late stage embryos and in the nervous system, ovaries, and the accessory glands of adult flies. RNAi was induced by injection of short (22 bp) beltless double-stranded RNAs into embryos or into adult flies. Embryonic RNAi altered cuticular phenotypes ranging from partially-formed to missing denticle belts (thus beltless) of the abdominal segments A2-A4. Embryonic beltless RNAi was lethal. Adult RNAi resulted in the shrinkage of the ovaries by half and reduced the number of eggs laid. We also examined Df(1)RK4 flies in which deletion removes 16 genes, including beltless. In some embryos, we observed cuticular abnormalities similar to our findings with beltless RNAi. After differentiating Df(1)RK4 embryos into those with visible denticle belts and those missing denticle belts, we assayed the presence of beltless mRNA; no beltless mRNA was detectable in embryos with missing denticle belts.
Conclusions: We have identified a developmentally important novel Drosophila gene, beltless, which has been characterized in loss-of-function studies using RNA interference. The putative beltless protein shares homologies with the C. elegans nose resistant to fluoxetine (NRF) NRF-6 gene, as well as with several uncharacterized C. elegans and Drosophila melanogaster genes, some with prominent acyltransferase domains. Future studies should elucidate the role and mechanism of action of beltless during Drosophila development and in adults, including in the adult nervous system.
Figures

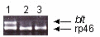


References
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

