Locating sequence on FPC maps and selecting a minimal tiling path
- PMID: 12915486
- PMCID: PMC403717
- DOI: 10.1101/gr.1068603
Locating sequence on FPC maps and selecting a minimal tiling path
Abstract
This study discusses three software tools, the first two aid in integrating sequence with an FPC physical map and the third automatically selects a minimal tiling path given genomic draft sequence and BAC end sequences. The first tool, FSD (FPC Simulated Digest), takes a sequenced clone and adds it back to the map based on a fingerprint generated by an in silico digest of the clone. This allows verification of sequenced clone positions and the integration of sequenced clones that were not originally part of the FPC map. The second tool, BSS (Blast Some Sequence), takes a query sequence and positions it on the map based on sequence associated with the clones in the map. BSS has multiple uses as follows: (1) When the query is a file of marker sequences, they can be added as electronic markers. (2) When the query is draft sequence, the results of BSS can be used to close gaps in a sequenced clone or the physical map. (3) When the query is a sequenced clone and the target is BAC end sequences, one may select the next clone for sequencing using both sequence comparison results and map location. (4) When the query is whole-genome draft sequence and the target is BAC end sequences, the results can be used to select many clones for a minimal tiling path at once. The third tool, pickMTP, automates the majority of this last usage of BSS. Results are presented using the rice FPC map, BAC end sequences, and whole-genome shotgun from Syngenta.
Figures
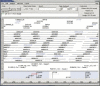
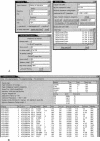




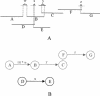
References
-
- Aho, A., Hopcroft, J., and Ullman, J. 1983. Data structures and algorithms. pp. 203–208. Addison-Wesley, Reading, MA.
-
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 769–815. - PubMed
-
- Bentley, D.R., Deloukas, P., Dunham, A., French, L., Gregory, S.G., Humphray, S.J., Mungall, A.J., Ross, M.T., Carter, N.P., Dunham, I., et al. 2001. The physical maps for sequencing human chromosomes 1, 6, 9, 10, 13, 20 and X. Nature 409: 942–943. - PubMed
WEB SITE REFERENCES
-
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; Smit, A.F.A. and Green, P., RepeatMasker.
-
- http://rgp.dna.affrc.go.jp/Publicdata.html; Japanese Rice Genome Research Program site for genetic markers and sequence.
-
- http://www.ensembl.org/Mus_musculus/; Ensembl Mouse Genome Server.
-
- http://www.genome.arizona.edu/fpc/rice; Rice Physical Mapping Home Page.
-
- http://www.genome.arizona.edu/fpc/rice/bss.html; Web-based BSS for rice.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
