Transcriptional response of lymphoblastoid cells to ionizing radiation
- PMID: 12915489
- PMCID: PMC403696
- DOI: 10.1101/gr.1240103
Transcriptional response of lymphoblastoid cells to ionizing radiation
Abstract
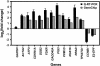
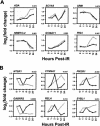
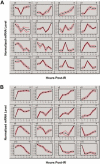
The effects of ionizing radiation (IR) on the temporal transcriptional response of lymphoblastoid cells were investigated in this study. We used oligonucleotide microarrays to assess mRNA levels of genes in lymphoblastoid cells at various time points within 24 h following gamma-irradiation. We identified 319 and 816 IR-responsive genes following 3 Gy and 10 Gy of IR exposure, respectively, with 126 genes in common between the two doses. A high percentage of IR-responsive genes are involved in the control of cell cycle, cell death, DNA repair, DNA metabolism, and RNA processing. We determined the temporal expression profiles of the IR-responsive genes and assessed effects of IR dose on this temporal pattern of expression. By combining dose-response data with temporal profiles of expression, we have identified sets of coordinately responding genes. Through a genomic approach, we characterized a set of genes that are implicated in cellular adaptation to IR stress. These findings will allow a better understanding of complex processes such as radiation-induced carcinogenesis and the development of biomarkers for radiation exposure.
Figures
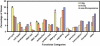

References
-
- Amundson, S.A., Bittner, M., Chen, Y., Trent, J., Meltzer, P., and Fornace Jr., A.J. 1999. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene 18: 3666–3672. - PubMed
-
- Amundson, S.A., Do, K.T., Shahab, S., Bittner, M., Meltzer, P., Trent, J., and Fornace Jr., A.J. 2000. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat. Res. 154: 342–346. - PubMed
-
- Amundson, S.A., Lee, R.A., Kock-Paiz, C.A., Bittner, M.L., Meltzer, P., Trent, J.M., and Fornace Jr., A.J. 2003. Differential responses of stress genes to low dose-rate γ irradiation. Mol. Cancer Res. 1: 445–452. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
