Targeting of the turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein
- PMID: 12915529
- PMCID: PMC187420
- DOI: 10.1128/jvi.77.17.9124-9135.2003
Targeting of the turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein
Abstract
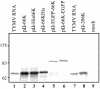
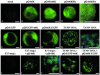
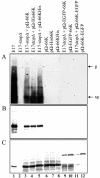
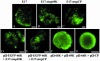
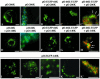
Turnip yellow mosaic virus (TYMV), a positive-strand RNA virus in the alphavirus-like superfamily, encodes two replication proteins, 140K and 66K, both being required for its RNA genome replication. The 140K protein contains domains indicative of methyltransferase, proteinase, and NTPase/helicase, and the 66K protein encompasses the RNA-dependent RNA polymerase domain. During viral infection, the 66K protein localizes to virus-induced chloroplastic membrane vesicles, which are closely associated with TYMV RNA replication. To investigate the determinants of its subcellular localization, the 66K protein was expressed in plant protoplasts from separate plasmids. Green fluorescent protein (GFP) fusion and immunofluorescence experiments demonstrated that the 66K protein displayed a cytoplasmic distribution when expressed individually but that it was relocated to the chloroplast periphery under conditions in which viral replication occurred. The 66K protein produced from an expression vector was functional in viral replication since it could transcomplement a defective replication template. Targeting of the 66K protein to the chloroplast envelope in the course of the viral infection appeared to be solely dependent on the expression of the 140K protein. Analysis of the subcellular localization of the 140K protein fused to GFP demonstrated that it is targeted to the chloroplast envelope in the absence of other viral factors and that it induces the clumping of the chloroplasts, one of the typical cytological effects of TYMV infection. These results suggests that the 140K protein is a key organizer of the assembly of the TYMV replication complexes and a major determinant for their chloroplastic localization and retention.
Figures

References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Wiley, New York, N.Y.
-
- Barends, S., H. H. Bink, S. H. van den Worm, C. W. Pleij, and B. Kraal. 2003. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell 112:123-129. - PubMed
-
- Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. - PubMed
-
- Bransom, K. L., S. E. Wallace, and T. W. Dreher. 1996. Identification of the cleavage site recognized by the turnip yellow mosaic virus protease. Virology 217:404-406. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

