Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells
- PMID: 12915536
- PMCID: PMC187424
- DOI: 10.1128/jvi.77.17.9204-9210.2003
Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells
Abstract
The hepatitis C virus (HCV) pandemic affects the health of more than 170 million people and is the major indication for orthotopic liver transplantations. Although the human liver is the primary site for HCV replication, it is not known whether extrahepatic tissues are also infected by the virus and whether nonprimate cells are permissive for RNA replication. Because HCV exists as a quasispecies, it is conceivable that a viral population may include variants that can replicate in different cell types and in other species. We have tested this hypothesis and found that subgenomic HCV RNAs can replicate in mouse hepatoma and nonhepatic human epithelial cells. Replicons isolated from these cell lines carry new mutations that could be involved in the control of tropism of the virus. Our results demonstrated that translation and RNA-directed RNA replication of HCV do not depend on hepatocyte or primate-specific factors. Moreover, our results could open the path for the development of animal models for HCV infection.
Figures

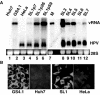

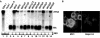
Comment in
-
Unexpected host range of hepatitis C virus replicons.Hepatology. 2004 Mar;39(3):835-8. doi: 10.1002/hep.20122. Hepatology. 2004. PMID: 14999704
References
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. - PubMed
-
- Boisvert, J., X. S. He, R. Cheung, E. B. Keeffe, T. Wright, and H. B. Greenberg. 2001. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J. Infect. Dis. 184:827-835. - PubMed
-
- Chang, T. T., K. C. Young, Y. J. Yang, H. Y. Lei, and H. L. Wu. 1996. Hepatitis C virus RNA in peripheral blood mononuclear cells: comparing acute and chronic hepatitis C virus infection. Hepatology 23:977-981. - PubMed
-
- Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. - PubMed
-
- Chun, T. W., J. S. Justement, P. Pandya, C. W. Hallahan, M. McLaughlin, S. Liu, L. A. Ehler, C. Kovacs, and A. S. Fauci. 2002. Relationship between the size of the human immunodeficiency virus type 1 (HIV-1) reservoir in peripheral blood CD4+ T cells and CD4+:CD8+ T cell ratios in aviremic HIV-1-infected individuals receiving long-term highly active antiretroviral therapy. J. Infect. Dis. 185:1672-1676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

