Complementation of a deletion in the rubella virus p150 nonstructural protein by the viral capsid protein
- PMID: 12915564
- PMCID: PMC187411
- DOI: 10.1128/jvi.77.17.9502-9510.2003
Complementation of a deletion in the rubella virus p150 nonstructural protein by the viral capsid protein
Abstract
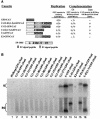
Rubella virus (RUB) replicons with an in-frame deletion of 507 nucleotides between two NotI sites in the P150 nonstructural protein (DeltaNotI) do not replicate (as detected by expression of a reporter gene encoded by the replicon) but can be amplified by wild-type helper virus (Tzeng et al., Virology 289:63-73, 2001). Surprisingly, virus with DeltaNotI was viable, and it was hypothesized that this was due to complementation of the NotI deletion by one of the virion structural protein genes. Introduction of the capsid (C) protein gene into DeltaNotI-containing replicons as an in-frame fusion with a reporter gene or cotransfection with both DeltaNotI replicons and RUB replicon or plasmid constructs containing the C gene resulted in replication of the DeltaNotI replicon, confirming the hypothesis that the C gene was the structural protein gene responsible for complementation and demonstrating that complementation could occur either in cis or in trans. Approximately the 5' one-third of the C gene was necessary for complementation. Mutations that prevented translation of the C protein while minimally disturbing the C gene sequence abrogated complementation, while synonymous codon mutations that changed the C gene sequence without affecting the amino acid sequence at the 5' end of the C gene had no effect on complementation, indicating that the C protein, not the C gene RNA, was the moiety responsible for complementation. Complementation occurred at a basic step in the virus replication cycle, because DeltaNotI replicons failed to accumulate detectable virus-specific RNA.
Figures


References
-
- Choi, I. R., and K. A. White. 2002. An RNA activator of subgenomic mRNA1 transcription in tomato bushy stunt virus. J. Biol. Chem. 277:3760-3766. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

