Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events
- PMID: 12915570
- PMCID: PMC187415
- DOI: 10.1128/jvi.77.17.9567-9577.2003
Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events
Abstract
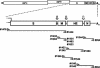
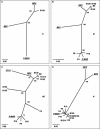
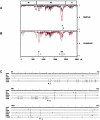
Toroviruses (family Coronaviridae, order Nidovirales) are enveloped, positive-stranded RNA viruses that have been implicated in enteric disease in cattle and possibly in humans. Despite their potential veterinary and clinical relevance, little is known about torovirus epidemiology and molecular genetics. Here, we present the first study into the diversity among toroviruses currently present in European swine and cattle herds. Comparative sequence analysis was performed focusing on the genes for the structural proteins S, M, HE, and N, with fecal specimens serving as sources of viral RNA. Sequence data published for animal and human torovirus variants were included. Four genotypes, displaying 30 to 40% divergence, were readily distinguished, exemplified by bovine torovirus (BToV) Breda, porcine torovirus (PToV) Markelo, equine torovirus Berne, and the putative human torovirus. The ungulate toroviruses apparently display host species preference. In phylogenetic analyses, all PToV variants clustered, while the recent European BToVs mostly resembled the New World BToV variant Breda, identified 19 years ago. However, we found ample evidence for recurring intertypic recombination. All newly characterized BToV variants seem to have arisen from a genetic exchange, during which the 3' end of the HE gene, the N gene, and the 3' nontranslated region of a Breda virus-like parent had been swapped for those of PToV. Moreover, some PToV and BToV variants carried chimeric HE genes, which apparently resulted from recombination events involving hitherto unknown toroviruses. From these observations, the existence of two additional torovirus genotypes can be inferred. Toroviruses may be even more promiscuous than their closest relatives, the coronaviruses and arteriviruses.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

