Point mutations in exon I of the herpes simplex virus putative terminase subunit, UL15, indicate that the most conserved residues are essential for cleavage and packaging
- PMID: 12915573
- PMCID: PMC187393
- DOI: 10.1128/jvi.77.17.9613-9621.2003
Point mutations in exon I of the herpes simplex virus putative terminase subunit, UL15, indicate that the most conserved residues are essential for cleavage and packaging
Abstract
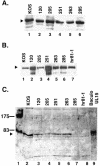
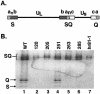
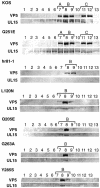
The herpes simplex virus UL15 and UL28 genes are believed to encode two subunits of the terminase involved in cleavage and packaging of viral genomes. Analysis of the UL15 protein sequence and its herpesvirus homologues revealed the presence of 20 conserved regions. Twelve of the twenty regions conserved among herpesviruses are also conserved in terminases from DNA bacteriophage. Point mutations in UL15 were designed in four conserved regions: L120N (CR1), Q205E (CR2), Q251E (CR3), G263A (CR3), and Y285S (CR4). Transfection experiments indicated that each mutant gene could produce stable UL15 protein at wild-type levels; however, only one mutant (Q251E) was able to complement the UL15-null virus. Each mutation was introduced into the viral genome by marker transfer, and all mutants except Q251E were unable to form plaques on Vero cells. Furthermore, failure to form plaques on Vero cells correlated with a defect in cleavage and packaging. Immunofluorescence experiments indicated that in cells infected with all mutant viruses the UL15 protein could be detected and was found to localize to replication compartments. Although wild-type and mutant Q251E were able to produce A, B, and C capsids, the rest of the mutants were only able to produce B capsids, a finding consistent with their defects in cleavage and packaging. In addition, all mutant UL15 proteins retained their ability to interact with B capsids. Therefore, amino acid residues 120, 205, 263, and 285 are essential for the cleavage and packaging process rather than for association with capsids or localization to replication compartments.
Figures



References
-
- Abbotts, A. P., V. G. Preston, M. Hughes, A. H. Patel, and N. D. Stow. 2000. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol. 81:2999-3009. - PubMed
-
- al-Kobaisi, M. F., F. J. Rizon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. J. Virol. 180:380-388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

