Fatty acid biosynthesis in Mycobacterium tuberculosis: lateral gene transfer, adaptive evolution, and gene duplication
- PMID: 12917487
- PMCID: PMC193559
- DOI: 10.1073/pnas.1737230100
Fatty acid biosynthesis in Mycobacterium tuberculosis: lateral gene transfer, adaptive evolution, and gene duplication
Abstract
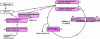
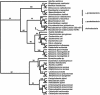
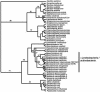
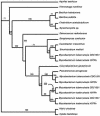
Mycobacterium tuberculosis is a high GC Gram-positive member of the actinobacteria. The mycobacterial cell wall is composed of a complex assortment of lipids and is the interface between the bacterium and its environment. The biosynthesis of fatty acids plays an essential role in the formation of cell wall components, in particular mycolic acids, which have been targeted by many of the drugs used to treat M. tuberculosis infection. M. tuberculosis has approximately 250 genes involved in fatty acid metabolism, a much higher proportion than in any other organism. In silico methods have been used to compare the genome of M. tuberculosis CDC1551 to a database of 58 complete bacterial genomes. The resulting alignments were scanned for genes specifically involved in fatty acid biosynthetic pathway I. Phylogenetic analysis of these alignments was used to investigate horizontal gene transfer, gene duplication, and adaptive evolution. It was found that of the eight gene families examined, five of the phylogenies reconstructed suggest that the actinobacteria have a closer relationship with the alpha-proteobacteria than expected. This is either due to either an ancient transfer of genes or deep paralogy and subsequent retention of the genes in unrelated lineages. Additionally, adaptive evolution and gene duplication have been an influence in the evolution of the pathway. This study provides a key insight into how M. tuberculosis has developed its unique fatty acid synthetic abilities.
Figures
References
-
- World Health Organization (2001) Stop TB Annual Report WHO/CDS/STB/2002.17 (W.H.O., Geneva).
-
- Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, et al. (1998) Nature 393, 537-544. - PubMed
-
- Valway, S. E., Sanchez, M. P., Shinnick, T. F., Orme, I., Agerton, T., Hoy, D., Jones, J. S., Westmoreland, H. & Onorato, I. M. (1998) N. Engl. J. Med. 338, 633-639. - PubMed
-
- Betts, J. C., Dodson, P., Quan, S., Lewis, A. P., Thomas, P. J., Duncan, K. & McAdam, R. A. (2000) Microbiology 146, 3205-3216. - PubMed
-
- Wolf, Y. I., Rogozin, I. B., Grishin, N. V. & Koonin, E. V. (2002) Trends Genet. 18, 472-479. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

