An integrated haplotype map of the human major histocompatibility complex
- PMID: 12920676
- PMCID: PMC1180682
- DOI: 10.1086/378101
An integrated haplotype map of the human major histocompatibility complex
Abstract
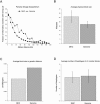
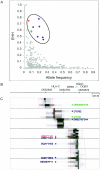
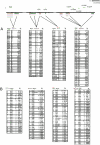
Numerous studies have clearly indicated a role for the major histocompatibility complex (MHC) in susceptibility to autoimmune diseases. Such studies have focused on the genetic variation of a small number of classical human-leukocyte-antigen (HLA) genes in the region. Although these genes represent good candidates, given their immunological roles, linkage disequilibrium (LD) surrounding these genes has made it difficult to rule out neighboring genes, many with immune function, as influencing disease susceptibility. It is likely that a comprehensive analysis of the patterns of LD and variation, by using a high-density map of single-nucleotide polymorphisms (SNPs), would enable a greater understanding of the nature of the observed associations, as well as lead to the identification of causal variation. We present herein an initial analysis of this region, using 201 SNPs, nine classical HLA loci, two TAP genes, and 18 microsatellites. This analysis suggests that LD and variation in the MHC, aside from the classical HLA loci, are essentially no different from those in the rest of the genome. Furthermore, these data show that multi-SNP haplotypes will likely be a valuable means for refining association signals in this region.
Figures

References
Electronic-Database Information
-
- Coriell Institute, http://locus.umdnj.edu/ccr/
-
- IDRG, http://www-genome.wi.mit.edu/mpg/idrg/projects/hla.html (for supplementary “Materials and Methods” information and pairwise D′ analysis for 201 reliable, polymorphic SNP assays in 18 multigenerational European CEPH families)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SLE, MS, IDDM, thyroid disease, and autoimmune diabetes)
References
-
- Allcock RJ, Atrazhev AM, Beck S, de Jong PJ, Elliott JF, Forbes S, Halls K, Horton R, Osoegawa K, Rogers J, Sawcer S, Todd JA, Trowsdale J, Wang Y, Williams S (2002) The MHC haplotype project: a resource for HLA-linked association studies. Tissue Antigens 59:520–521 - PubMed
-
- Alper CA, Awdeh Z, Yunis EJ (1992) Conserved, extended MHC haplotypes. Exp Clin Immunogenet 9:58–71 - PubMed
-
- Barcellos LF, Oksenberg JR, Green AJ, Bucher P, Rimmler JB, Schmidt S, Garcia ME Lincoln RR, Pericak-Vance MA, Haines JL, Hauser SL, Multiple Sclerosis Genetic Group (2002) Genetic basis for clinical expression in multiple sclerosis. Brain 125:150–158 - PubMed
-
- Beck S, Trowsdale J (2000) The human major histocompatability complex: lessons from the DNA sequence. Annu Rev Genomics Hum Genet 1:117–137 - PubMed
-
- Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fildes N, Bugawan TL, Erlich HA, Klitz W (1992) Polymorphism, recombination, and linkage disequilibrium within the HLA class II region. J Immunol 148:249–258 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

