Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior
- PMID: 12923056
- PMCID: PMC196259
- DOI: 10.1101/gad.1103303
Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior
Abstract
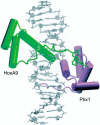
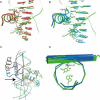
The HOX/HOM superfamily of homeodomain proteins controls cell fate and segmental embryonic patterning by a mechanism that is conserved in all metazoans. The linear arrangement of the Hox genes on the chromosome correlates with the spatial distribution of HOX protein expression along the anterior-posterior axis of the embryo. Most HOX proteins bind DNA cooperatively with members of the PBC family of TALE-type homeodomain proteins, which includes human Pbx1. Cooperative DNA binding between HOX and PBC proteins requires a residue N-terminal to the HOX homeodomain termed the hexapeptide, which differs significantly in sequence between anterior- and posterior-regulating HOX proteins. We report here the 1.9-A-resolution structure of a posterior HOX protein, HoxA9, complexed with Pbx1 and DNA, which reveals that the posterior Hox hexapeptide adopts an altered conformation as compared with that seen in previously determined anterior HOX/PBC structures. The additional nonspecific interactions and altered DNA conformation in this structure account for the stronger DNA-binding affinity and altered specificity observed for posterior HOX proteins when compared with anterior HOX proteins. DNA-binding studies of wild-type and mutant HoxA9 and HoxB1 show residues in the N-terminal arm of the homeodomains are critical for proper DNA sequence recognition despite lack of direct contact by these residues to the DNA bases. These results help shed light on the mechanism of transcriptional regulation by HOX proteins and show how DNA-binding proteins may use indirect contacts to determine sequence specificity.
Figures



References
-
- Billeter, M., Qian, Y.Q., Otting, G., Muller, M., Gehring, W., and Wuthrich, K. 1993. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain–DNA complex. J. Mol. Biol. 234: 1084–1093. - PubMed
-
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 (Pt 5): 905–921. - PubMed
-
- Chan, S.K., Jaffe, L., Capovilla, M., Botas, J., and Mann, R.S. 1994. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 78: 603–615. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
