Control of SXT integration and excision
- PMID: 12923077
- PMCID: PMC181012
- DOI: 10.1128/JB.185.17.5045-5054.2003
Control of SXT integration and excision
Abstract
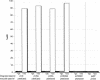
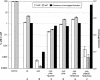
The Vibrio cholerae SXT element is a conjugative self-transmissible chromosomally integrating element that encodes resistance to multiple antibiotics. SXT integrates in a site-specific fashion at prfC and excises from the chromosome to form a circular but nonreplicative extrachromosomal form. Both chromosomal integration and excision depend on an SXT-encoded recombinase, Int. Here we found that Int is necessary and sufficient for SXT integration and that int expression in recipient cells requires the SXT activators SetC and SetD. Although no xis-like gene was annotated in the SXT genome, Int was not sufficient to mediate efficient SXT chromosomal excision. We identified a novel SXT Xis that seems to function as a recombination directionality factor (RDF), facilitating SXT excision and inhibiting SXT integration. Although unrelated to any previously characterized RDF, Xis is similar to five hypothetical proteins that together may constitute a new family of RDFs. Using real-time quantitative PCR assays to study SXT excision from the chromosome, we determined that while SXT excision is required for SXT transfer, the percentage of cells containing an excised circular SXT does not appear to be a major factor limiting SXT transfer; i.e., we found that most cells harboring an excised circular SXT molecule do not act as SXT donors. In the absence of prfC, SXT integrated into several secondary attachment sites but preferentially into the 5' end of pntB. SXT excision and transfer from a donor containing pntB::SXT were reduced, suggesting that the SXT integration site may also influence the element's transmissibility.
Figures

References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Azaro, M. A., and A. Landy. 2002. The integration/excision cycle of lambda and other bacteriophages, p. 117-148. In N. L. Craig, R. Craigie, M. Gellert, and A. Lambovitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
-
- Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

