Cyclic AMP receptor protein-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism
- PMID: 12923087
- PMCID: PMC180988
- DOI: 10.1128/JB.185.17.5148-5157.2003
Cyclic AMP receptor protein-dependent activation of the Escherichia coli acsP2 promoter by a synergistic class III mechanism
Abstract
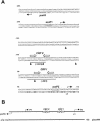
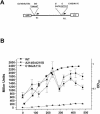
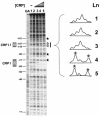
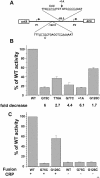
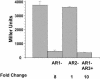
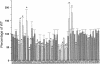
The cyclic AMP receptor protein (CRP) activates transcription of the Escherichia coli acs gene, which encodes an acetate-scavenging enzyme required for fitness during periods of carbon starvation. Two promoters direct transcription of acs, the distal acsP1 and the proximal acsP2. In this study, we demonstrated that acsP2 can function as the major promoter and showed by in vitro studies that CRP facilitates transcription by "focusing" RNA polymerase to acsP2. We proposed that CRP activates transcription from acsP2 by a synergistic class III mechanism. Consistent with this proposal, we showed that CRP binds two sites, CRP I and CRP II. Induction of acs expression absolutely required CRP I, while optimal expression required both CRP I and CRP II. The locations of these DNA sites for CRP (centered at positions -69.5 and -122.5, respectively) suggest that CRP interacts with RNA polymerase through class I interactions. In support of this hypothesis, we demonstrated that acs transcription requires the surfaces of CRP and the C-terminal domain of the alpha subunit of RNA polymerase holoenzyme (alpha-CTD), which is known to participate in class I interactions: activating region 1 of CRP and the 287, 265, and 261 determinants of the alpha-CTD. Other surface-exposed residues in the alpha-CTD contributed to acs transcription, suggesting that the alpha-CTD may interact with at least one protein other than CRP.
Figures
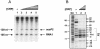
References
-
- Benoff, B., H. Yang, C. L. Lawson, G. Parkinson, J. Liu, E. Blatter, Y. W. Ebright, H. M. Berman, and R. H. Ebright. 2002. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science 297:1562-1566. - PubMed
-
- Berg, P. 1956. Acyl adenylates: an enzymatic mechanism of acetate activation. J. Biol. Chem. 222:991-1013. - PubMed
-
- Brown, T. D. K., M. C. Jones-Mortimer, and H. L. Kornberg. 1977. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102:327-336. - PubMed
-
- Browning, D. F., C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. W. Busby. 2002. Independent regulation of the divergent Escherichia coli nrfA and acsP1 promoters by a nucleoprotein assembly at a shared regualtory region. Mol. Microbiol. 43:687-701. - PubMed
-
- Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

