The SopEPhi phage integrates into the ssrA gene of Salmonella enterica serovar Typhimurium A36 and is closely related to the Fels-2 prophage
- PMID: 12923091
- PMCID: PMC181011
- DOI: 10.1128/JB.185.17.5182-5191.2003
The SopEPhi phage integrates into the ssrA gene of Salmonella enterica serovar Typhimurium A36 and is closely related to the Fels-2 prophage
Abstract
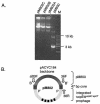
Salmonella spp. are enteropathogenic gram-negative bacteria that use a large array of virulence factors to colonize the host, manipulate host cells, and resist the host's defense mechanisms. Even closely related Salmonella strains have different repertoires of virulence factors. Bacteriophages contribute substantially to this diversity. There is increasing evidence that the reassortment of virulence factor repertoires by converting phages like the GIFSY phages and SopEPhi may represent an important mechanism in the adaptation of Salmonella spp. to specific hosts and to the emergence of new epidemic strains. Here, we have analyzed in more detail SopEPhi, a P2-like phage from Salmonella enterica serovar Typhimurium DT204 that encodes the virulence factor SopE. We have cloned and characterized the attachment site (att) of SopEPhi and found that its 47-bp core sequence overlaps the 3' terminus of the ssrA gene of serovar Typhimurium. Furthermore, we have demonstrated integration of SopEPhi into the cloned attB site of serovar Typhimurium A36. Sequence analysis of the plasmid-borne prophage revealed that SopEPhi is closely related to (60 to 100% identity over 80% of the genome) but clearly distinct from the Fels-2 prophage of serovar Typhimurium LT2 and from P2-like phages in the serovar Typhi CT18 genome. Our results demonstrate that there is considerable variation among the P2-like phages present in closely related Salmonella spp.
Figures






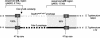

Similar articles
-
Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella.Mol Microbiol. 2001 Jan;39(2):260-71. doi: 10.1046/j.1365-2958.2001.02234.x. Mol Microbiol. 2001. PMID: 11136448
-
Variability in occurrence of multiple prophage genes in Salmonella Typhimurium strains isolated in Slovak Republic.FEMS Microbiol Lett. 2007 May;270(2):237-44. doi: 10.1111/j.1574-6968.2007.00674.x. Epub 2007 Mar 13. FEMS Microbiol Lett. 2007. PMID: 17355601
-
Discrimination within phenotypically closely related definitive types of Salmonella enterica serovar typhimurium by the multiple amplification of phage locus typing technique.J Clin Microbiol. 2005 Apr;43(4):1604-11. doi: 10.1128/JCM.43.4.1604-1611.2005. J Clin Microbiol. 2005. PMID: 15814973 Free PMC article.
-
Bacteriophage-encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium.Infect Genet Evol. 2005 Jan;5(1):1-9. doi: 10.1016/j.meegid.2004.07.004. Infect Genet Evol. 2005. PMID: 15567133 Review.
-
Salmonella phages and prophages--genomics and practical aspects.Methods Mol Biol. 2007;394:133-75. doi: 10.1007/978-1-59745-512-1_9. Methods Mol Biol. 2007. PMID: 18363236 Review.
Cited by
-
Transposition of the heat-stable toxin astA gene into a gifsy-2-related prophage of Salmonella enterica serovar Abortusovis.J Bacteriol. 2004 Jul;186(14):4568-74. doi: 10.1128/JB.186.14.4568-4574.2004. J Bacteriol. 2004. PMID: 15231789 Free PMC article.
-
A novel phage element of Salmonella enterica serovar Enteritidis P125109 contributes to accelerated type III secretion system 2-dependent early inflammation kinetics in a mouse colitis model.Infect Immun. 2012 Sep;80(9):3236-46. doi: 10.1128/IAI.00180-12. Epub 2012 Jul 2. Infect Immun. 2012. PMID: 22753379 Free PMC article.
-
Salmonella-Based Biorodenticides: Past Applications and Current Contradictions.Int J Mol Sci. 2022 Nov 23;23(23):14595. doi: 10.3390/ijms232314595. Int J Mol Sci. 2022. PMID: 36498920 Free PMC article. Review.
-
Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion.Microbiol Mol Biol Rev. 2004 Sep;68(3):560-602, table of contents. doi: 10.1128/MMBR.68.3.560-602.2004. Microbiol Mol Biol Rev. 2004. PMID: 15353570 Free PMC article. Review.
-
Use of the lambda Red recombinase system to produce recombinant prophages carrying antibiotic resistance genes.BMC Mol Biol. 2006 Sep 19;7:31. doi: 10.1186/1471-2199-7-31. BMC Mol Biol. 2006. PMID: 16984631 Free PMC article.
References
-
- Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-490. - PubMed
-
- Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. - PubMed
-
- Brussow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-223. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

