Polynucleotide phosphorylase-deficient mutants of Pseudomonas putida
- PMID: 12923102
- PMCID: PMC180990
- DOI: 10.1128/JB.185.17.5279-5286.2003
Polynucleotide phosphorylase-deficient mutants of Pseudomonas putida
Abstract
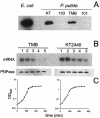
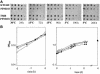
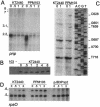
In bacteria, polynucleotide phosphorylase (PNPase) is one of the main exonucleolytic activities involved in RNA turnover and is widely conserved. In spite of this, PNPase does not seem to be essential for growth if the organisms are not subjected to special conditions, such as low temperature. We identified the PNPase-encoding gene (pnp) of Pseudomonas putida and constructed deletion mutants that did not exhibit cold sensitivity. In addition, we found that the transcription pattern of pnp upon cold shock in P. putida was markedly different from that in Escherichia coli. It thus appears that pnp expression control and the physiological roles in the cold may be different in different bacterial species.
Figures

Similar articles
-
Transcriptional and post-transcriptional control of polynucleotide phosphorylase during cold acclimation in Escherichia coli.Mol Microbiol. 2000 Jun;36(6):1470-80. doi: 10.1046/j.1365-2958.2000.01971.x. Mol Microbiol. 2000. PMID: 10931296
-
Polynucleotide phosphorylase hinders mRNA degradation upon ribosomal protein S1 overexpression in Escherichia coli.RNA. 2008 Nov;14(11):2417-29. doi: 10.1261/rna.1123908. Epub 2008 Sep 29. RNA. 2008. PMID: 18824515 Free PMC article.
-
RNase activity of polynucleotide phosphorylase is critical at low temperature in Escherichia coli and is complemented by RNase II.J Bacteriol. 2008 Sep;190(17):5924-33. doi: 10.1128/JB.00500-08. Epub 2008 Jul 7. J Bacteriol. 2008. PMID: 18606734 Free PMC article.
-
Regulation and functions of bacterial PNPase.Wiley Interdiscip Rev RNA. 2016 Mar-Apr;7(2):241-58. doi: 10.1002/wrna.1328. Epub 2016 Jan 11. Wiley Interdiscip Rev RNA. 2016. PMID: 26750178 Review.
-
Human polynucleotide phosphorylase (hPNPase(old-35)): an evolutionary conserved gene with an expanding repertoire of RNA degradation functions.Oncogene. 2011 Apr 14;30(15):1733-43. doi: 10.1038/onc.2010.572. Epub 2010 Dec 13. Oncogene. 2011. PMID: 21151174 Free PMC article. Review.
Cited by
-
Long-term survival of Campylobacter jejuni at low temperatures is dependent on polynucleotide phosphorylase activity.Appl Environ Microbiol. 2009 Dec;75(23):7310-8. doi: 10.1128/AEM.01366-09. Epub 2009 Oct 2. Appl Environ Microbiol. 2009. PMID: 19801468 Free PMC article.
-
Transient expression of βC1 protein differentially regulates host genes related to stress response, chloroplast and mitochondrial functions.Virol J. 2010 Dec 30;7:373. doi: 10.1186/1743-422X-7-373. Virol J. 2010. PMID: 21192819 Free PMC article.
-
Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis.J Bacteriol. 2008 May;190(10):3467-74. doi: 10.1128/JB.00075-08. Epub 2008 Mar 14. J Bacteriol. 2008. PMID: 18344363 Free PMC article.
-
Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica.Infect Immun. 2006 Feb;74(2):1243-54. doi: 10.1128/IAI.74.2.1243-1254.2006. Infect Immun. 2006. PMID: 16428774 Free PMC article.
-
The world of ribonucleases from pseudomonads: a short trip through the main features and singularities.Microb Biotechnol. 2021 Nov;14(6):2316-2333. doi: 10.1111/1751-7915.13890. Epub 2021 Aug 24. Microb Biotechnol. 2021. PMID: 34427985 Free PMC article. Review.
References
-
- Bagdasarian, M., R. Lurz, B. Ruckert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. - PubMed
-
- Baggi, G., D. Catelani, C. Sorlini, and V. Treccani. 1987. Microbial degradation of methylbenzenes: metabolism of 1,2,4-trimethylbenzene by a Pseudomonas putida. Ann. Microbiol. 32:45.
-
- Beran, R. K., and R. W. Simons. 2001. Cold-temperature induction of Escherichia coli polynucleotide phosphorylase occurs by reversal of its autoregulation. Mol. Microbiol. 39:112-125. - PubMed
-
- Bestetti, G., P. Di Gennaro, E. Galli, B. Leoni, Pellizzoni, Sello, and Bianchi. 1994. Bioconversion of substityute naphtalenes to the corresponding salicylic acid. Appl. Microbiol. Biotechnol. 40:791-793.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

