siRNA function in RNAi: a chemical modification analysis
- PMID: 12923253
- PMCID: PMC1370469
- DOI: 10.1261/rna.5103703
siRNA function in RNAi: a chemical modification analysis
Abstract
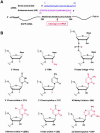
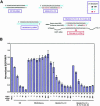
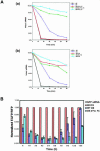
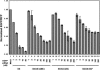
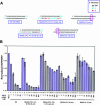
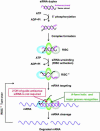
Various chemical modifications were created in short-interfering RNAs (siRNAs) to determine the biochemical properties required for RNA interference (RNAi). Remarkably, modifications at the 2'-position of pentose sugars in siRNAs showed the 2'-OHs were not required for RNAi, indicating that RNAi machinery does not require the 2'-OH for recognition of siRNAs and catalytic ribonuclease activity of RNA-induced silencing complexes (RISCs) does not involve the 2'-OH of guide antisense RNA. In addition, 2' modifications predicted to stabilize siRNA increased the persistence of RNAi as compared with wild-type siRNAs. RNAi was also induced with chemical modifications that stabilized interactions between A-U base pairs, demonstrating that these types of modifications may enhance mRNA targeting efficiency in allele-specific RNAi. Modifications altering the structure of the A-form major groove of antisense siRNA-mRNA duplexes abolished RNAi, suggesting that the major groove of these duplexes was required for recognition by activated RISC*. Comparative analysis of the stability and RNAi activities of chemically modified single-stranded antisense RNA and duplex siRNA suggested that some catalytic mechanism(s) other than siRNA stability were linked to RNAi efficiency. Modified or mismatched ribonucleotides incorporated at internal positions in the 5' or 3' half of the siRNA duplex, as defined by the antisense strand, indicated that the integrity of the 5' and not the 3' half of the siRNA structure was important for RNAi, highlighting the asymmetric nature of siRNA recognition for initiation of unwinding. Collectively, this study defines the mechanisms of RNAi in human cells and provides new rules for designing effective and stable siRNAs for RNAi-mediated gene-silencing applications.
Figures

References
-
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. - PubMed
-
- Chiu, Y.L. and Rana, T.M. 2002. RNAi in human cells: Basic structural and functional features of small interfering RNA. Mol. Cell 10: 549–561. - PubMed
-
- Cogoni, C. and Macino, G. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
