Zeptomole detection of a viral nucleic acid using a target-activated ribozyme
- PMID: 12923255
- PMCID: PMC1370471
- DOI: 10.1261/rna.5760703
Zeptomole detection of a viral nucleic acid using a target-activated ribozyme
Abstract
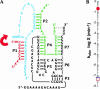
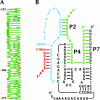
We describe a strategy for the ultra-sensitive detection of nucleic acids using "half" ribozymes that are devoid of catalytic activity unless completed by a trans-acting target nucleic acid. The half-ribozyme concept was initially demonstrated using a construct derived from a multiple turnover Class I ligase. Iterative RNA selection was carried out to evolve this half-ribozyme into one activated by a conserved sequence present in the hepatitis C virus (HCV) genome. Following sequence optimization of substrate RNAs, this HCV-activated half-ribozyme displayed a maximal turnover rate of 69 min(-1) (pH 8.3) and was induced in rate by approximately 2.6 x 10(9)-fold by the HCV target. It detected the HCV target oligonucleotide in the zeptomole range (6700 molecules), a sensitivity of detection roughly 2.6 x 10(6)-fold greater than that previously demonstrated by oligonucleotide-activated ribozymes, and one that is sufficient for molecular diagnostic applications.
Figures




Similar articles
-
High-throughput ribozyme-based assays for detection of viral nucleic acids.Chem Biol. 2004 Jun;11(6):807-15. doi: 10.1016/j.chembiol.2004.03.029. Chem Biol. 2004. PMID: 15217614
-
Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons.FEBS J. 2005 Nov;272(22):5910-22. doi: 10.1111/j.1742-4658.2005.04986.x. FEBS J. 2005. PMID: 16279954
-
[Determination of the in vitro antiviral activity of an engineered M1GS ribozyme that targets to the core gene of hepatitis C virus].Wei Sheng Wu Xue Bao. 2013 Aug 4;53(8):875-81. Wei Sheng Wu Xue Bao. 2013. PMID: 24341280 Chinese.
-
Activity of HDV ribozymes to trans-cleave HCV RNA.World J Gastroenterol. 2002 Aug;8(4):694-8. doi: 10.3748/wjg.v8.i4.694. World J Gastroenterol. 2002. PMID: 12174380 Free PMC article.
-
Ribozymes of the hepatitis delta virus: recent findings on their structure, mechanism of catalysis and possible applications.Acta Biochim Pol. 2001;48(2):409-18. Acta Biochim Pol. 2001. PMID: 11732611 Review.
Cited by
-
Direct selection of trans-acting ligase ribozymes by in vitro compartmentalization.RNA. 2005 Oct;11(10):1555-62. doi: 10.1261/rna.2121705. Epub 2005 Aug 30. RNA. 2005. PMID: 16131588 Free PMC article.
-
An RNA polymerase ribozyme that synthesizes its own ancestor.Proc Natl Acad Sci U S A. 2020 Feb 11;117(6):2906-2913. doi: 10.1073/pnas.1914282117. Epub 2020 Jan 27. Proc Natl Acad Sci U S A. 2020. PMID: 31988127 Free PMC article.
-
Topological rearrangement yields structural stabilization and interhelical distance constraints in the Kin.46 self-phosphorylating ribozyme.RNA. 2006 Dec;12(12):2118-25. doi: 10.1261/rna.173506. Epub 2006 Oct 26. RNA. 2006. PMID: 17068208 Free PMC article.
-
Assembly and activation of a kinase ribozyme.RNA. 2010 Dec;16(12):2349-59. doi: 10.1261/rna.2302810. Epub 2010 Oct 8. RNA. 2010. PMID: 20935068 Free PMC article.
-
Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods.Nucleic Acids Res. 2011 Sep 1;39(16):e110. doi: 10.1093/nar/gkr504. Epub 2011 Jun 21. Nucleic Acids Res. 2011. PMID: 21693555 Free PMC article.
References
-
- Bartel, D.P. and Szostak, J.W. 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418. - PubMed
-
- Bergman, N.H., Johnston, W.K., and Bartel, D.P. 2000. Kinetic framework for ligation by an efficient RNA ligase ribozyme. Biochemistry 39: 3115–3123. - PubMed
-
- Breaker, R.R. 2002. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 13: 31–39. - PubMed
-
- Burke, D.H., Ozerova, N.D., and Nilsen-Hamilton, M. 2002. Allosteric hammerhead ribozyme TRAPs. Biochemistry 41: 6588–6594. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
