The potential for gene repair via triple helix formation
- PMID: 12925687
- PMCID: PMC171401
- DOI: 10.1172/JCI19552
The potential for gene repair via triple helix formation
Abstract
Triplex-forming oligonucleotides (TFOs) can bind to polypurine/polypyrimidine regions in DNA in a sequence-specific manner. The specificity of this binding raises the possibility of using triplex formation for directed genome modification, with the ultimate goal of repairing genetic defects in human cells. Several studies have demonstrated that treatment of mammalian cells with TFOs can provoke DNA repair and recombination, in a manner that can be exploited to introduce desired sequence changes. This review will summarize recent advances in this field while also highlighting major obstacles that remain to be overcome before the application of triplex technology to therapeutic gene repair can be achieved.
Figures

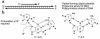
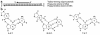
References
-
- Felsenfeld G, Davies DR, Rich A. Formation of a three stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024.
-
- Thuong NT, Helene C. Sequence specific recognition and modification of double helical DNA by oligonucleotides. Angewandte Chemie. Intl. Ed. Eng. 1993;32:666–690.
-
- Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu. Rev. Biochem. 1995;64:65–95. - PubMed
-
- Manor H, Rao BS, Martin RG. Abundance and degree of dispersion of genomic d(GA)n.d(TC)n sequences. J. Mol. Evol. 1988;27:96–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

