Visualization of chromatin domains created by the gypsy insulator of Drosophila
- PMID: 12925706
- PMCID: PMC2173808
- DOI: 10.1083/jcb.200305013
Visualization of chromatin domains created by the gypsy insulator of Drosophila
Abstract
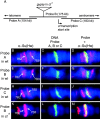
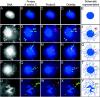
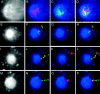
Insulators might regulate gene expression by establishing and maintaining the organization of the chromatin fiber within the nucleus. Biochemical fractionation and in situ high salt extraction of lysed cells show that two known protein components of the gypsy insulator are present in the nuclear matrix. Using FISH with DNA probes located between two endogenous Su(Hw) binding sites, we show that the intervening DNA is arranged in a loop, with the two insulators located at the base. Mutations in insulator proteins, subjecting the cells to a brief heat shock, or destruction of the nuclear matrix lead to disruption of the loop. Insertion of an additional gypsy insulator in the center of the loop results in the formation of paired loops through the attachment of the inserted sequences to the nuclear matrix. These results suggest that the gypsy insulator might establish higher-order domains of chromatin structure and regulate nuclear organization by tethering the DNA to the nuclear matrix and creating chromatin loops.
Figures


References
-
- Bell, A.C., A.G. West, and G. Felsenfeld. 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 291:447–450. - PubMed
-
- Berger, S.L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142–148. - PubMed
-
- Cai, H.N., and P. Shen. 2001. Effects of cis arrangement of chromatin insulators on ennhancer-blocking activity. Science. 291:493–495. - PubMed
-
- Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins, P. van Sluis, M.C. Hermus, R. van Asperen, K. Boon, P.A. Voute, et al. 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 291:1289–1292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

