Schizosacchromyces pombe Dpb2 binds to origin DNA early in S phase and is required for chromosomal DNA replication
- PMID: 12925774
- PMCID: PMC181578
- DOI: 10.1091/mbc.e03-02-0088
Schizosacchromyces pombe Dpb2 binds to origin DNA early in S phase and is required for chromosomal DNA replication
Abstract
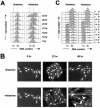
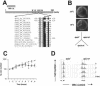
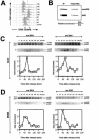
Genetic evidence suggests that DNA polymerase epsilon (Pol epsilon) has a noncatalytic essential role during the early stages of DNA replication initiation. Herein, we report the cloning and characterization of the second largest subunit of Pol epsilon in fission yeast, called Dpb2. We demonstrate that Dpb2 is essential for cell viability and that a temperature-sensitive mutant of dpb2 arrests with a 1C DNA content, suggesting that Dpb2 is required for initiation of DNA replication. Using a chromatin immunoprecipitation assay, we show that Dpb2, binds preferentially to origin DNA at the beginning of S phase. We also show that the C terminus of Pol epsilon associates with origin DNA at the same time as Dpb2. We conclude that Dpb2 is an essential protein required for an early step in DNA replication. We propose that the primary function of Dpb2 is to facilitate assembly of the replicative complex at the start of S phase. These conclusions are based on the novel cell cycle arrest phenotype of the dpb2 mutant, on the previously uncharacterized binding of Dpb2 to replication origins, and on the observation that the essential function of Pol epsilon is not dependent on its DNA synthesis activity.
Figures



References
-
- Bell, S.P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

