Chaperone-mediated in vitro assembly of Polyomavirus capsids
- PMID: 12928495
- PMCID: PMC193586
- DOI: 10.1073/pnas.1832245100
Chaperone-mediated in vitro assembly of Polyomavirus capsids
Abstract
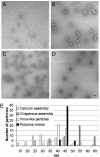
The polyomavirus coat protein viral protein 1 (VP1) has the intrinsic ability to self-assemble in vitro into polymorphic capsid-like structures on addition of calcium. In contrast, polyomavirus assembly in vivo is rigorously controlled, such that virions of uniform size are formed only in the cell nucleus. During viral infection, the 72 kDa cellular chaperone heat shock cognate protein (hsc70) binds VP1 posttranslation and colocalizes with VP1 to the nucleus, thereby suggesting a role for approximately 70-kDa heat shock protein (hsp70) family chaperones in regulating the quality and location of capsid assembly. We found that, after expression of recombinant VP1 in Escherichia coli, the prokaryotic hsp70 chaperone DnaK copurified with the VP1 C-terminal domain that links pentamers in an assembled capsid. When stably bound to VP1, DnaK inhibited in vitro assembly induced by calcium. However, in the presence of ATP, the hsp70 chaperone system comprised of DnaK, DnaJ, and GrpE assembled VP1 into uniform capsids without requiring calcium. Chaperone-mediated assembly was similarly catalyzed by the eukaryotic hsc70 protein, in combination with the J-domain function of the simian virus 40 large T-antigen protein. Thus, polyomavirus capsid assembly can be recapitulated with high-fidelity in vitro using either prokaryotic or eukaryotic hsp70 chaperone systems, thereby supporting a role for cellular chaperones in the in vivo regulation of virion assembly.
Figures



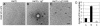
References
-
- Liddington, R. C., Yan, Y., Moulai, J., Sahli, R., Benjamin, T. L. & Harrison, S. C. (1991) Nature 354, 278-284. - PubMed
-
- Griffith, J. P., Griffith, D. L., Rayment, I., Murakami, W. T. & Caspar, D. L. (1992) Nature 355, 652-654. - PubMed
-
- Garcea, R. L., Salunke, D. M. & Caspar, D. L. (1987) Nature 329, 86-87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

