Reconstitution of recombination-dependent DNA synthesis in herpes simplex virus 1
- PMID: 12928502
- PMCID: PMC193539
- DOI: 10.1073/pnas.1534569100
Reconstitution of recombination-dependent DNA synthesis in herpes simplex virus 1
Abstract
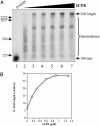
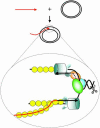
The repair of double-strand DNA breaks by homologous recombination is essential for the maintenance of genome stability. In herpes simplex virus 1, double-strand DNA breaks may arise as a consequence of replication fork collapse at sites of oxidative damage, which is known to be induced upon viral infection. Double-strand DNA breaks are also generated by cleavage of viral a sequences by endonuclease G during genome isomerization. We have reconstituted a system using purified proteins in which strand invasion is coupled with DNA synthesis. In this system, the viral single-strand DNA-binding protein promotes assimilation of single-stranded DNA into a homologous supercoiled plasmid, resulting in the formation of a displacement loop. The 3' terminus of the invading DNA serves as a primer for long-chain DNA synthesis promoted by the viral DNA replication proteins, including the polymerase and helicase-primase. Efficient extension of the invading primer also requires a DNA-relaxing enzyme (eukaryotic topoisomerase I or DNA gyrase). The viral polymerase by itself is insufficient for DNA synthesis, and a DNA-relaxing enzyme cannot substitute for the viral helicase-primase. The viral single-strand DNA-binding protein, in addition to its role in the invasion process, is also required for long-chain DNA synthesis. Form X, a topologically distinct, positively supercoiled form of displacement-loop, does not serve as a template for DNA synthesis. These observations support a model in which recombination and replication contribute toward maintaining viral genomic stability by repairing double-strand breaks. They also account for the extensive branching observed during viral replication in vivo.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

