Hierarchical model of gene regulation by transforming growth factor beta
- PMID: 12930890
- PMCID: PMC193550
- DOI: 10.1073/pnas.1834070100
Hierarchical model of gene regulation by transforming growth factor beta
Abstract
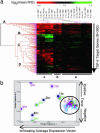
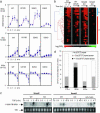
Transforming growth factor betas (TGF-betas) regulate key aspects of embryonic development and major human diseases. Although Smad2, Smad3, and extracellular signal-regulated kinase (ERK) mitogen-activated protein kinases (MAPKs) have been proposed as key mediators in TGF-beta signaling, their functional specificities and interactivity in controlling transcriptional programs in different cell types and (patho)physiological contexts are not known. We investigated expression profiles of genes controlled by TGF-beta in fibroblasts with ablations of Smad2, Smad3, and ERK MAPK. Our results suggest that Smad3 is the essential mediator of TGF-beta signaling and directly activates genes encoding regulators of transcription and signal transducers through Smad3/Smad4 DNA-binding motif repeats that are characteristic for immediate-early target genes of TGF-beta but absent in intermediate target genes. In contrast, Smad2 and ERK predominantly transmodulated regulation of both immediate-early and intermediate genes by TGF-beta/Smad3. These results suggest a previously uncharacterized hierarchical model of gene regulation by TGF-beta in which TGF-beta causes direct activation by Smad3 of cascades of regulators of transcription and signaling that are transmodulated by Smad2 and/or ERK.
Figures
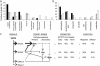
References
-
- Derynck, R., Akhurst, R. J. & Balmain, A. (2001) Nat. Genet. 29, 117-129. - PubMed
-
- Roberts, A. B. (2002) Cytokine Growth Factor Rev. 13, 3-5. - PubMed
-
- Massague, J. (2000) Nat. Rev. Mol. Cell Biol. 1, 169-178. - PubMed
-
- Dennler, S., Huet, S. & Gauthier, J. M. (1999) Oncogene 18, 1643-1648. - PubMed
-
- Yagi, K., Goto, D., Hamamoto, T., Takenoshita, S., Kato, M. & Miyazono, K. (1999) J. Biol. Chem. 274, 703-709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

