The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA
- PMID: 12930902
- PMCID: PMC193545
- DOI: 10.1073/pnas.1434308100
The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA
Abstract
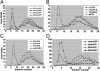
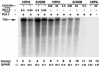
The linkage of sister chromatids after DNA replication ensures the faithful inheritance of chromosomes by daughter cells. In budding yeast, the establishment of sister chromatid cohesion requires Ctf8, Dcc1, and Ctf18, a homologue of the p140 subunit of the replication factor C (RFC). In this report we demonstrate that in 293T cells, Flag-tagged Ctf18 forms a seven-subunit cohesion-RFC complex comprised of Ctf18, Dcc1, Ctf8, RFCp40, RFCp38, RFCp37, and RFCp36 (Ctf18-RFC). We demonstrate that a stoichiometric heteroheptameric Ctf18-RFC complex can be assembled by coexpressing the seven proteins in baculovirus-infected insect cells. In addition, the two other stable subcomplexes were formed, which include a pentameric complex comprised of Ctf18, RFCp40, RFCp38, RFCp37, and RFCp36 and a dimeric Dcc1-Ctf8. Both the five- and seven-subunit Ctf18-RFC complexes bind to single-stranded and primed DNAs and possess weak ATPase activity that is stimulated by the addition of primed DNA and proliferating cell nuclear antigen (PCNA). These complexes catalyzed the ATP-dependent loading of PCNA onto primed and gapped DNA but not onto double-stranded nicked or single-stranded circular DNAs. Consistent with these observations, both Ctf18-RFC complexes substituted for the replicative RFC in the PCNA-dependent DNA polymerase delta-catalyzed DNA replication reaction. These results support a model in which sister chromatid cohesion is linked to DNA replication.
Figures
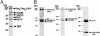



References
-
- Nasmyth, K. (2001) Annu. Rev. Genet. 35, 673-745. - PubMed
-
- Cahill, D. P., Lengauer, C., Yu, J., Riggins, G. J., Willson, J. K., Markowitz, S. D., Kinzler, K. W. & Vogelstein, B. (1998) Nature 392, 300-303. - PubMed
-
- Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature 386, 623-627. - PubMed
-
- Wolstenholme, J. & Angell, R. R. (2000) Chromosoma 109, 435-438. - PubMed
-
- Hirano, T. (2000) Annu. Rev. Biochem. 69, 115-144. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

