Differences in replication of a DNA template containing an ethyl phosphotriester by T4 DNA polymerase and Escherichia coli DNA polymerase I
- PMID: 12930945
- PMCID: PMC212818
- DOI: 10.1093/nar/gkg722
Differences in replication of a DNA template containing an ethyl phosphotriester by T4 DNA polymerase and Escherichia coli DNA polymerase I
Abstract
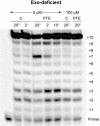
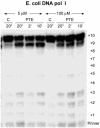
A DNA template containing a single ethyl phosphotriester was replicated in vitro by the bacteriophage T4 DNA polymerase and by Escherichia coli DNA polymerase I (DNA pol I). Escherichia coli DNA pol I bypassed the lesion efficiently, but partial inhibition was observed for T4 DNA polymerase. The replication block produced by the ethyl phosphotriester was increased at low dNTP concentrations and for a mutant T4 DNA polymerase with an antimutator phenotype, increased proofreading activity, and reduced ability to bind DNA in the polymerase active center. These observations support a model in which an ethyl phosphotriester impedes primer elongation by T4 DNA polymerase by decreasing formation of the ternary DNA polymerase-DNA-dNTP complex. When primer elongation is not possible, proofreading becomes the favored reaction. Apparent futile cycles of nucleotide incorporation and proofreading, the idling reaction, were observed at the site of the lesion. The replication block was overcome by higher dNTP concentrations. Thus, ethyl phosphotriesters may be tolerated in vivo by the up-regulation of dNTP biosynthesis that occurs during the cellular checkpoint response to blocked DNA replication forks.
Figures



Similar articles
-
The nucleotide analog 2-aminopurine as a spectroscopic probe of nucleotide incorporation by the Klenow fragment of Escherichia coli polymerase I and bacteriophage T4 DNA polymerase.Biochemistry. 1995 Jul 18;34(28):9185-92. doi: 10.1021/bi00028a031. Biochemistry. 1995. PMID: 7619819
-
DNA polymerase mutagenic bypass and proofreading of endogenous DNA lesions.Mutat Res. 1999 Mar 8;424(1-2):221-36. doi: 10.1016/s0027-5107(99)00021-4. Mutat Res. 1999. PMID: 10064863
-
Processive proofreading is intrinsic to T4 DNA polymerase.J Biol Chem. 1992 Jul 15;267(20):14157-66. J Biol Chem. 1992. PMID: 1629215
-
Understanding DNA replication by the bacteriophage T4 replisome.J Biol Chem. 2017 Nov 10;292(45):18434-18442. doi: 10.1074/jbc.R117.811208. Epub 2017 Sep 25. J Biol Chem. 2017. PMID: 28972188 Free PMC article. Review.
-
Regulation of DNA polymerase exonucleolytic proofreading activity: studies of bacteriophage T4 "antimutator" DNA polymerases.Genetics. 1998 Apr;148(4):1551-7. doi: 10.1093/genetics/148.4.1551. Genetics. 1998. PMID: 9560374 Free PMC article. Review. No abstract available.
Cited by
-
John W. (Jan) Drake: A Biochemical View of a Geneticist Par Excellence.Genetics. 2020 Dec;216(4):827-836. doi: 10.1534/genetics.120.303813. Genetics. 2020. PMID: 33268388 Free PMC article.
-
Engineering processive DNA polymerases with maximum benefit at minimum cost.Front Microbiol. 2014 Aug 4;5:380. doi: 10.3389/fmicb.2014.00380. eCollection 2014. Front Microbiol. 2014. PMID: 25136334 Free PMC article.
-
Methyl DNA Phosphate Adduct Formation in Rats Treated Chronically with 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone and Enantiomers of Its Metabolite 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanol.Chem Res Toxicol. 2018 Jan 16;31(1):48-57. doi: 10.1021/acs.chemrestox.7b00281. Epub 2017 Nov 30. Chem Res Toxicol. 2018. PMID: 29131934 Free PMC article.
-
Detection of phosphodiester adducts formed by the reaction of benzo[a]pyrene diol epoxide with 2'-deoxynucleotides using collision-induced dissociation electrospray ionization tandem mass spectrometry.Nucleic Acids Res. 2007;35(15):5014-27. doi: 10.1093/nar/gkm526. Epub 2007 Jul 18. Nucleic Acids Res. 2007. PMID: 17636312 Free PMC article.
-
Mass Spectrometric Quantitation of Pyridyloxobutyl DNA Phosphate Adducts in Rats Chronically Treated with N'-Nitrosonornicotine.Chem Res Toxicol. 2019 Apr 15;32(4):773-783. doi: 10.1021/acs.chemrestox.9b00007. Epub 2019 Feb 26. Chem Res Toxicol. 2019. PMID: 30740971 Free PMC article.
References
-
- Snow E.T., Foote,R.S. and Mitra,S. (1984) Base-pairing properties of O6-methylguanine in template DNA during in vitro DNA replication. J. Biol. Chem., 259, 8095–8100. - PubMed
-
- Singer B., Chavez,F., Goodman,M.F., Essigmann,J.M. and Dosanjh M.K. (1989) Effect of 3′ flanking neighbors on kinetics of pairing of dCTP or dTTP opposite O6-methylguanine in a defined primed oligonucleotide when Escherichia coli DNA polymerase I is used. Proc. Natl Acad. Sci. USA, 86, 8271–8274. - PMC - PubMed
-
- Reha-Krantz L.J., Nonay,R.L., Day,R.S.,III and Wilson,S.H. (1996) Replication of O6-methylguanine-containing DNA by repair and replicative DNA polymerases. J. Biol. Chem., 271, 20088–20095. - PubMed
-
- Miller P.S., Chandrasegaran,S., Dow,D.L., Pulford,M. and Kan,L.S. (1982) Synthesis and template properties of an ethyl phosphotriester modified decadeoxyribonucleotide. Biochemistry, 21, 5468–5474. - PubMed

