Construction and analysis of cells lacking the HMGA gene family
- PMID: 12930952
- PMCID: PMC212792
- DOI: 10.1093/nar/gkg684
Construction and analysis of cells lacking the HMGA gene family
Abstract
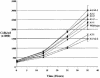
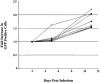
The high mobility group A (HMGA) family of non-histone chromosomal proteins is encoded by two related genes, HMGA1 and HMGA2. HMGA proteins are architectural transcription factors that have been found to regulate the transcription of a large number of genes. They are also some of the most commonly dysregulated genes in human neoplasias, highlighting a role in growth control. HMGA1 and HMGA2 have also been found to stimulate retroviral integration in vitro. In this study, we have cloned chicken HMGA1, and used the chicken DT40 B-cell lymphoma line to generate cells lacking HMGA1, HMGA2 and both in combination. We tested these lines for effects on cellular growth, gene control and retroviral integration. Surprisingly, we found that the HMGA gene family is dispensable for growth in DT40 cells, and that there is no apparent defect in retroviral integration in the absence of HMGA1 or HMGA2. We also analyzed the activity of approximately 4000 chicken genes, but found no significant changes. We conclude that HMGA proteins are not strictly required for growth control or retroviral integration in DT40 cells and may well be redundant with other factors.
Figures




References
-
- Goodwin G.H., Sanders,C. and Johns,E.W. (1973) A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur. J. Biochem., 38, 14–19. - PubMed
-
- Grosschedl R., Giese,K. and Pagel,J. (1994) HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet., 10, 94–100. - PubMed
-
- Johnson K.R., Disney,J.E., Wyatt,C.R. and Reeves,R. (1990) Expression of mRNAs encoding mammalian chromosomal proteins HMG-I and HMG-Y during cellular proliferation. Exp. Cell Res., 187, 69–76. - PubMed
-
- Nagpal S., Ghosn,C., DiSepio,D., Molina,Y., Sutter,M., Klein,E.S. and Chandraratna,R.A. (1999) Retinoid-dependent recruitment of a histone H1 displacement activity by retinoic acid receptor. J. Biol. Chem., 274, 22563–22568. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions

