3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures
- PMID: 12930962
- PMCID: PMC212791
- DOI: 10.1093/nar/gkg680
3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures
Abstract
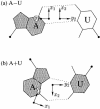
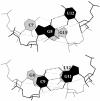
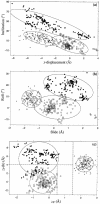
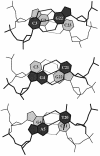
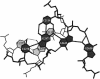
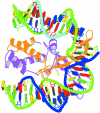
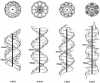
We present a comprehensive software package, 3DNA, for the analysis, reconstruction and visualization of three-dimensional nucleic acid structures. Starting from a coordinate file in Protein Data Bank (PDB) format, 3DNA can handle antiparallel and parallel double helices, single-stranded structures, triplexes, quadruplexes and other complex tertiary folding motifs found in both DNA and RNA structures. The analysis routines identify and categorize all base interactions and classify the double helical character of appropriate base pair steps. The program makes use of a recently recommended reference frame for the description of nucleic acid base pair geometry and a rigorous matrix-based scheme to calculate local conformational parameters and rebuild the structure from these parameters. The rebuilding routines produce rectangular block representations of nucleic acids as well as full atomic models with the sugar-phosphate backbone and publication quality 'standardized' base stacking diagrams. Utilities are provided to locate the base pairs and helical regions in a structure and to reorient structures for effective visualization. Regular helical models based on X-ray diffraction measurements of various repeating sequences can also be generated within the program.
Figures

References
-
- Watson J.D. and Crick,F.H.C. (1953) Genetical implications of the structure of deoxyribonucleic acid. Nature, 171, 964–967. - PubMed
-
- Saenger W. (1984) Principles of Nucleic Acid Structure. Springer-Verlag, New York, NY, Ch. 6.
-
- Kang C.H., Zhang,X., Ratliff,R., Moyzis,R. and Rich,A. (1992) Crystal structure of four-stranded Oxytricha telomeric DNA. Nature, 356, 126–131. - PubMed
-
- Smith F.W. and Feigon,J. (1992) Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature, 356, 164–168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

