Comparative analysis of the base biases at the gene terminal portions in seven eukaryote genomes
- PMID: 12930971
- PMCID: PMC212801
- DOI: 10.1093/nar/gkg701
Comparative analysis of the base biases at the gene terminal portions in seven eukaryote genomes
Abstract
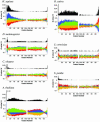
Adenine nucleotides have been found to appear preferentially in the regions after the initiation codons or before the termination codons of bacterial genes. Our previous experiments showed that AAA and AAT, the two most frequent second codons in Escherichia coli, significantly enhance translation efficiency. To determine whether such a characteristic feature of base frequencies exists in eukaryote genes, we performed a comparative analysis of the base biases at the gene terminal portions using the proteomes of seven eukaryotes. Here we show that the base appearance at the codon third positions of gene terminal regions is highly biased in eukaryote genomes, although the codon third positions are almost free from amino acid preference. The bias changes depending on its position in a gene, and is characteristic of each species. We also found that bias is most outstanding at the second codon, the codon after the initiation codon. NCN is preferred in every genome; in particular, GCG is strongly favored in human and plant genes. The presence of the bias implies that the base sequences at the second codon affect translation efficiency in eukaryotes as well as bacteria.
Figures

References
-
- Sato T., Terabe,M., Watanabe,H., Gojobori,T., Hori-Takemoto,C. and Miura,K. (2001) Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem., 129, 851–860. - PubMed
-
- Stenström C.M., Jin,H., Major,L.L., Tate,W.P. and Isaksson,L.A. (2001) Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene, 263, 273–284. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

