Parallel gene analysis with allele-specific padlock probes and tag microarrays
- PMID: 12930977
- PMCID: PMC212823
- DOI: 10.1093/nar/gng104
Parallel gene analysis with allele-specific padlock probes and tag microarrays
Abstract
Parallel, highly specific analysis methods are required to take advantage of the extensive information about DNA sequence variation and of expressed sequences. We present a scalable laboratory technique suitable to analyze numerous target sequences in multiplexed assays. Sets of padlock probes were applied to analyze single nucleotide variation directly in total genomic DNA or cDNA for parallel genotyping or gene expression analysis. All reacted probes were then co-amplified and identified by hybridization to a standard tag oligonucleotide array. The technique was illustrated by analyzing normal and pathogenic variation within the Wilson disease-related ATP7B gene, both at the level of DNA and RNA, using allele-specific padlock probes.
Figures

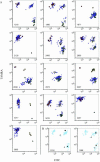
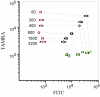
References
-
- Syvanen A.C. (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nature Rev. Genet., 2, 930–942. - PubMed
-
- Wang D.G., Fan,J.B., Siao,C.J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J. et al. (1998) Large-scale identification, mapping and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280, 1077–1082. - PubMed
-
- Patil N., Berno,A.J., Hinds,D.A., Barrett,W.A., Doshi,J.M., Hacker,C.R., Kautzer,C.R., Lee,D.H., Marjoribanks,C., McDonough,D.P. et al. (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science, 294, 1719–1723. - PubMed
-
- Oliphant A., Barker,D.L., Stuelpnagel,J.R. and Chee,M.S. (2002) BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques, 32, 56–61. - PubMed
-
- Yeakley J.M., Fan,J.B., Doucet,D., Luo,L., Wickham,E., Ye,Z., Chee,M.S. and Fu,X.D. (2002) Profiling alternative splicing on fiber-optic arrays. Nat. Biotechnol., 20, 353–358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

