Kinetic Outlier Detection (KOD) in real-time PCR
- PMID: 12930979
- PMCID: PMC212825
- DOI: 10.1093/nar/gng106
Kinetic Outlier Detection (KOD) in real-time PCR
Abstract
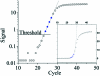
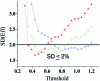
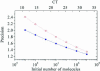
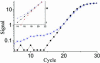
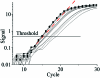
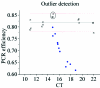
Real-time PCR is becoming the method of choice for precise quantification of minute amounts of nucleic acids. For proper comparison of samples, almost all quantification methods assume similar PCR efficiencies in the exponential phase of the reaction. However, inhibition of PCR is common when working with biological samples and may invalidate the assumed similarity of PCR efficiencies. Here we present a statistical method, Kinetic Outlier Detection (KOD), to detect samples with dissimilar efficiencies. KOD is based on a comparison of PCR efficiency, estimated from the amplification curve of a test sample, with the mean PCR efficiency of samples in a training set. KOD is demonstrated and validated on samples with the same initial number of template molecules, where PCR is inhibited to various degrees by elevated concentrations of dNTP; and in detection of cDNA samples with an aberrant ratio of two genes. Translating the dissimilarity in efficiency to quantity, KOD identifies outliers that differ by 1.3-1.9-fold in their quantity from normal samples with a P-value of 0.05. This precision is higher than the minimal 2-fold difference in number of DNA molecules that real-time PCR usually aims to detect. Thus, KOD may be a useful tool for outlier detection in real-time PCR.
Figures
References
-
- Livak K.J. (1997) ABI Prism 7700 Sequence Detection System, User Bulletin 2. PE Applied Biosystems.
-
- Gentle A., Anastasopoulos,F. and McBrien,N.A. (2001) High-resolution semi-quantitative real-time PCR without the use of a standard curve. Biotechniques, 31, 502, 504,–506, 508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

