Resistance to the peptidyl transferase inhibitor tiamulin caused by mutation of ribosomal protein l3
- PMID: 12936991
- PMCID: PMC182624
- DOI: 10.1128/AAC.47.9.2892-2896.2003
Resistance to the peptidyl transferase inhibitor tiamulin caused by mutation of ribosomal protein l3
Abstract
The antibiotic tiamulin targets the 50S subunit of the bacterial ribosome and interacts at the peptidyl transferase center. Tiamulin-resistant Escherichia coli mutants were isolated in order to elucidate mechanisms of resistance to the drug. No mutations in the rRNA were selected as resistance determinants using a strain expressing only a plasmid-encoded rRNA operon. Selection in a strain with all seven chromosomal rRNA operons yielded a mutant with an A445G mutation in the gene coding for ribosomal protein L3, resulting in an Asn149Asp alteration. Complementation experiments and sequencing of transductants demonstrate that the mutation is responsible for the resistance phenotype. Chemical footprinting experiments show a reduced binding of tiamulin to mutant ribosomes. It is inferred that the L3 mutation, which points into the peptidyl transferase cleft, causes tiamulin resistance by alteration of the drug-binding site. This is the first report of a mechanism of resistance to tiamulin unveiled in molecular detail.
Figures

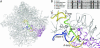

References
-
- Ban, N., P. Nissen, J. Hansen, P. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. - PubMed
-
- Douthwaite, S. R., T. Powers, J. Y. Lee, and H. F. Noller. 1989. Defining the structural requirements for a helix in 23 S ribosomal RNA that confers erythromycin resistance. J. Mol. Biol. 209:655-665. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

