Human mitochondrial C1-tetrahydrofolate synthase: gene structure, tissue distribution of the mRNA, and immunolocalization in Chinese hamster ovary calls
- PMID: 12937168
- PMCID: PMC1457088
- DOI: 10.1074/jbc.M304319200
Human mitochondrial C1-tetrahydrofolate synthase: gene structure, tissue distribution of the mRNA, and immunolocalization in Chinese hamster ovary calls
Abstract
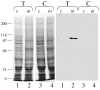
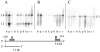
C1-tetrahydrofolate (THF) synthase is a trifunctional enzyme found in eukaryotes that contains the activities 10-formyl-THF synthetase, 5,10-methenyl-THF cyclohydrolase, and 5,10-methylene-THF dehydrogenase. The cytoplasmic isozyme of C1-THF synthase is well characterized in a number of mammals, including humans; but a mitochondrial isozyme has been previously identified only in the yeast Saccharomyces. Here, we report the identification and characterization of the human gene encoding a functional mitochondrial C1-THF synthase. The gene spans 236 kilobase pairs on chromosome 6 and consists of 28 exons plus one alternative exon. The gene encodes a protein of 978 amino acids, including an N-terminal mitochondrial targeting sequence. The mitochondrial isozyme is 61% identical to the human cytoplasmic isozyme. Expression of the gene was detected in most human tissues, but transcripts were highest in placenta, thymus, and brain. Two mRNAs were detected, a 3.6-kb transcript and a 1.1-kb transcript, and both transcripts were observed in varying ratios in each tissue. The shorter transcript results from an alternative splicing event, where exon 7 is spliced to exon 8a instead of exon 8. Exon 8a is derived from an exonized Alu sequence, sharing no homology with exon 8 of the long transcript, and encodes just 15 amino acids followed by a stop codon and a polyadenylation signal. This short transcript potentially encodes a bifunctional enzyme lacking 10-formyl-THF synthetase activity. Both transcripts initiate at the same 5'-site, 107 nucleotides up-stream of the ATG start codon. The full-length (2934 bp) cDNA fused to a C-terminal V5 epitope tag was expressed in Chinese hamster ovary cells. Immunoblots of subfractionated cells revealed a 107-kDa protein only in the mitochondrial fractions of these cells, confirming the mitochondrial localization of the protein. Yeast cells expressing the full-length human cDNA exhibited elevated 10-formyl-THF synthetase activity, confirming its identification as the human mitochondrial C1-THF synthase.
Figures


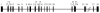

Similar articles
-
Enzymatic characterization of human mitochondrial C1-tetrahydrofolate synthase.Arch Biochem Biophys. 2005 Oct 15;442(2):196-205. doi: 10.1016/j.abb.2005.08.007. Epub 2005 Aug 30. Arch Biochem Biophys. 2005. PMID: 16171773
-
A novel mitochondrial C1-tetrahydrofolate synthetase is upregulated in human colon adenocarcinoma.Biochem Biophys Res Commun. 2004 Feb 27;315(1):204-11. doi: 10.1016/j.bbrc.2004.01.035. Biochem Biophys Res Commun. 2004. PMID: 15013446
-
Human mitochondrial C1-tetrahydrofolate synthase: submitochondrial localization of the full-length enzyme and characterization of a short isoform.Arch Biochem Biophys. 2009 Jan 1;481(1):86-93. doi: 10.1016/j.abb.2008.10.028. Epub 2008 Oct 29. Arch Biochem Biophys. 2009. PMID: 18996079 Free PMC article.
-
Primary structure of a folate-dependent trifunctional enzyme from Spodoptera frugiperda.Biochim Biophys Acta. 1995 Mar 14;1261(1):129-33. doi: 10.1016/0167-4781(95)00004-z. Biochim Biophys Acta. 1995. PMID: 7893749
-
Mammalian mitochondrial methylenetetrahydrofolate dehydrogenase-cyclohydrolase derived from a trifunctional methylenetetrahydrofolate dehydrogenase-cyclohydrolase-synthetase.Arch Biochem Biophys. 2002 Jul 1;403(1):145-8. doi: 10.1016/S0003-9861(02)00203-5. Arch Biochem Biophys. 2002. PMID: 12061812
Cited by
-
Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells.Cell Death Dis. 2013 Oct 24;4(10):e877. doi: 10.1038/cddis.2013.393. Cell Death Dis. 2013. PMID: 24157871 Free PMC article.
-
A global characterization and identification of multifunctional enzymes.PLoS One. 2012;7(6):e38979. doi: 10.1371/journal.pone.0038979. Epub 2012 Jun 18. PLoS One. 2012. PMID: 22723914 Free PMC article.
-
Folate Metabolism in Hepatocellular Carcinoma. What Do We Know So Far?Technol Cancer Res Treat. 2022 Jan-Dec;21:15330338221144446. doi: 10.1177/15330338221144446. Technol Cancer Res Treat. 2022. PMID: 36503290 Free PMC article. Review.
-
Molecular structure of a 5,10-methylenetetrahydrofolate dehydrogenase from the silkworm Bombyx mori.FEBS Open Bio. 2019 Feb 26;9(4):618-628. doi: 10.1002/2211-5463.12595. eCollection 2019 Apr. FEBS Open Bio. 2019. PMID: 30984537 Free PMC article.
-
Polymorphisms in MTHFD1 Gene and Susceptibility to Neural Tube Defects: A Case-Control Study in a Chinese Han Population with Relatively Low Folate Levels.Med Sci Monit. 2015 Sep 4;21:2630-7. doi: 10.12659/MSM.895155. Med Sci Monit. 2015. PMID: 26343515 Free PMC article.
References
-
- Appling DR. FASEB J. 1991;5:2645–2651. - PubMed
-
- Paukert JL, Williams GR, Rabinowitz JC. Biochem. Biophys. Res. Comm. 1977;77:147–154. - PubMed
-
- Shannon KW, Rabinowitz JC. J. Biol. Chem. 1986;261:12266–12271. - PubMed
-
- Paukert JL, Straus LDA, Rabinowitz JC. J. Biol. Chem. 1976;251:5104–5111. - PubMed
-
- Tan LUL, MacKenzie RE. Can. J. Biochem. 1979;57:806–812. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

