Structure of the bacteriophage T4 DNA adenine methyltransferase
- PMID: 12937411
- PMCID: PMC4030375
- DOI: 10.1038/nsb973
Structure of the bacteriophage T4 DNA adenine methyltransferase
Abstract
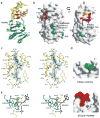
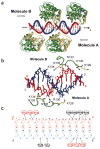
DNA-adenine methylation at certain GATC sites plays a pivotal role in bacterial and phage gene expression as well as bacterial virulence. We report here the crystal structures of the bacteriophage T4Dam DNA adenine methyltransferase (MTase) in a binary complex with the methyl-donor product S-adenosyl-L-homocysteine (AdoHcy) and in a ternary complex with a synthetic 12-bp DNA duplex and AdoHcy. T4Dam contains two domains: a seven-stranded catalytic domain that harbors the binding site for AdoHcy and a DNA binding domain consisting of a five-helix bundle and a beta-hairpin that is conserved in the family of GATC-related MTase orthologs. Unexpectedly, the sequence-specific T4Dam bound to DNA in a nonspecific mode that contained two Dam monomers per synthetic duplex, even though the DNA contains a single GATC site. The ternary structure provides a rare snapshot of an enzyme poised for linear diffusion along the DNA.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures

Similar articles
-
Bacteriophage T4Dam (DNA-(adenine-N6)-methyltransferase): evidence for two distinct stages of methylation under single turnover conditions.J Biol Chem. 2003 Oct 24;278(43):41749-55. doi: 10.1074/jbc.M306397200. Epub 2003 Jul 30. J Biol Chem. 2003. PMID: 12893823
-
Bacteriophage T4Dam DNA-(adenine-N(6))-methyltransferase. Comparison of pre-steady state and single turnover methylation of 40-mer duplexes containing two (un)modified target sites.J Biol Chem. 2004 Nov 26;279(48):50012-8. doi: 10.1074/jbc.M409786200. Epub 2004 Sep 16. J Biol Chem. 2004. PMID: 15375160
-
Study of bacteriophage T4-encoded Dam DNA (adenine-N6)-methyltransferase binding with substrates by rapid laser UV cross-linking.J Biol Chem. 2007 Sep 7;282(36):26067-76. doi: 10.1074/jbc.M700866200. Epub 2007 Jul 13. J Biol Chem. 2007. PMID: 17630395
-
[Molecular enzymology of phage T4 Dam DNA-methyltransferase].Mol Biol (Mosk). 2004 Sep-Oct;38(5):869-85. Mol Biol (Mosk). 2004. PMID: 15554189 Review. Russian.
-
Bacteriophage T2Dam and T4Dam DNA-[N6-adenine]-methyltransferases.Prog Nucleic Acid Res Mol Biol. 2004;77:67-126. doi: 10.1016/S0079-6603(04)77003-8. Prog Nucleic Acid Res Mol Biol. 2004. PMID: 15196891 Review. No abstract available.
Cited by
-
Mutations within the catalytic motif of DNA adenine methyltransferase (Dam) of Aeromonas hydrophila cause the virulence of the Dam-overproducing strain to revert to that of the wild-type phenotype.Infect Immun. 2006 Oct;74(10):5763-72. doi: 10.1128/IAI.00994-06. Infect Immun. 2006. PMID: 16988254 Free PMC article.
-
Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions.Biol Chem. 2004 May;385(5):373-9. doi: 10.1515/BC.2004.041. Biol Chem. 2004. PMID: 15195996 Free PMC article.
-
Structural and functional insights into the molecular mechanism of rRNA m6A methyltransferase RlmJ.Nucleic Acids Res. 2013 Nov;41(20):9537-48. doi: 10.1093/nar/gkt719. Epub 2013 Aug 13. Nucleic Acids Res. 2013. PMID: 23945937 Free PMC article.
-
Escherichia coli DNA adenine methyltransferase: the structural basis of processive catalysis and indirect read-out.J Biol Chem. 2009 Jul 3;284(27):18390-400. doi: 10.1074/jbc.M109.005876. Epub 2009 May 5. J Biol Chem. 2009. PMID: 19419959 Free PMC article.
-
Methyltransferases acquired by lactococcal 936-type phage provide protection against restriction endonuclease activity.BMC Genomics. 2014 Oct 1;15(1):831. doi: 10.1186/1471-2164-15-831. BMC Genomics. 2014. PMID: 25269955 Free PMC article.
References
-
- Lacks S, Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977;114:153–168. - PubMed
-
- Hattman S, Brooks JE, Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978;126:367–380. - PubMed
-
- Bale A, d’Alarcao M, Marinus MG. Characterization of DNA adenine methylation mutants of Escherichia coli K12. Mutat Res. 1979;59:157–165. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
