New reporter gene-based replication assay reveals exchangeability of replication factors of porcine circovirus types 1 and 2
- PMID: 12941898
- PMCID: PMC224580
- DOI: 10.1128/jvi.77.18.9885-9893.2003
New reporter gene-based replication assay reveals exchangeability of replication factors of porcine circovirus types 1 and 2
Abstract
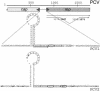
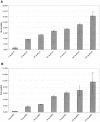
Two types of porcine circovirus (PCV), which differ in their pathogenicity, are known. PCV type 2 (PCV2) is the etiological agent of postweaning multisystemic wasting syndrome in swine, while PCV1 has not yet been linked to a disease. Corroborating earlier observations in PCV1, transcript mapping revealed that the rep gene of PCV2 encodes two products, the full-length protein Rep and the spliced version Rep' and that the simultaneous expression of Rep and Rep' proteins is essential for initiation of replication of PCV2. The interchangeability of the replication factors of PCV1 and PCV2 was examined. The rep gene products of PCV2 were not only able to bind the PCV2 origin but also the origin of PCV1 and vice versa. To investigate the competence of the Rep/Rep' proteins to initiate replication at the heterologous origin, a new replication assay was developed. It measures the expression of a luc reporter gene present on a plasmid carrying the origin of the investigated replicon. Replication is initiated by expression of the appendant replicase from a second plasmid and results in replication of the origin plasmid coupled with an increase in the Luc activity. Using this method to compare replication of PCV1 and PCV2 in cell culture, it was shown that the Rep/Rep' protein of PCV2 initiated replication at the origin of PCV1, as did the reciprocal combination. Our results indicate that the cis- and trans-acting replication factors of the two viruses are functionally exchangeable.
Figures

Similar articles
-
Molecular biology of Porcine circovirus: analyses of gene expression and viral replication.Vet Microbiol. 2004 Feb 4;98(2):81-8. doi: 10.1016/j.vetmic.2003.10.014. Vet Microbiol. 2004. PMID: 14741119 Review.
-
PCV2 replicase transcripts in infected porcine kidney (PK15) cells.Virus Genes. 2002 Dec;25(3):323-8. doi: 10.1023/a:1020940411267. Virus Genes. 2002. PMID: 12881643
-
Transcriptional analysis of porcine circovirus type 2.Virology. 2003 Jan 5;305(1):168-80. doi: 10.1006/viro.2002.1733. Virology. 2003. PMID: 12504550
-
Functional exchangeability of the nuclear localization signal (NLS) of capsid protein between PCV1 and PCV2 in vitro: Implications for the role of NLS in viral replication.Virol J. 2011 Jul 6;8:341. doi: 10.1186/1743-422X-8-341. Virol J. 2011. PMID: 21733152 Free PMC article.
-
Porcine circoviruses--small but powerful.Virus Res. 2009 Aug;143(2):177-83. doi: 10.1016/j.virusres.2009.02.009. Epub 2009 Feb 26. Virus Res. 2009. PMID: 19647885 Review.
Cited by
-
A survey on porcine circovirus type 2 infection and phylogenetic analysis of its ORF2 gene in Hangzhou, Zhejiang Province, China.J Zhejiang Univ Sci B. 2008 Feb;9(2):148-53. doi: 10.1631/jzus.B0710395. J Zhejiang Univ Sci B. 2008. PMID: 18257137 Free PMC article.
-
Identification of Putative ORF5 Protein of Porcine Circovirus Type 2 and Functional Analysis of GFP-Fused ORF5 Protein.PLoS One. 2015 Jun 2;10(6):e0127859. doi: 10.1371/journal.pone.0127859. eCollection 2015. PLoS One. 2015. PMID: 26035722 Free PMC article.
-
Functional analysis of cis- and trans-acting replication factors of porcine circovirus type 1.J Virol. 2007 Jun;81(11):5696-704. doi: 10.1128/JVI.02420-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360750 Free PMC article.
-
The porcine circovirus type 1 capsid gene promoter improves antigen expression and immunogenicity in a HIV-1 plasmid vaccine.Virol J. 2011 Feb 7;8:51. doi: 10.1186/1743-422X-8-51. Virol J. 2011. PMID: 21299896 Free PMC article.
-
Computational based design and tracking of synthetic variants of Porcine circovirus reveal relations between silent genomic information and viral fitness.Sci Rep. 2021 May 19;11(1):10620. doi: 10.1038/s41598-021-89918-6. Sci Rep. 2021. PMID: 34012100 Free PMC article.
References
-
- Cheung, A. K. 2003. Transcriptional analysis of porcine circovirus type 2. Virology 305:168-180. - PubMed
-
- Koonin, E. V., and T. V. Ilyina. 1993. Computer-assisted dissection of rolling circle DNA replication. Biosystems 30:241-268. - PubMed
-
- Leung, A. Y., M. Chan, S. C. Tang, R. Liang, and Y. L. Kwong. 2002. Real-time quantitative analysis of polyoma BK viremia and viruria in renal allograft recipients. J. Virol. Methods 103:51-56. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

