Estimation of population bottlenecks during systemic movement of tobacco mosaic virus in tobacco plants
- PMID: 12941900
- PMCID: PMC224588
- DOI: 10.1128/jvi.77.18.9906-9911.2003
Estimation of population bottlenecks during systemic movement of tobacco mosaic virus in tobacco plants
Abstract
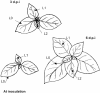
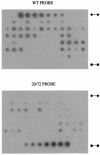
More often than not, analyses of virus evolution have considered that virus populations are so large that evolution can be explained by purely deterministic models. However, virus populations could have much smaller effective numbers than the huge reported census numbers, and random genetic drift could be important in virus evolution. A reason for this would be population bottlenecks during the virus life cycle. Here we report a quantitative estimate of population bottlenecks during the systemic colonization of tobacco leaves by Tobacco mosaic virus (TMV). Our analysis is based on the experimental estimation of the frequency of different genotypes of TMV in the inoculated leaf, and in systemically infected leaves, of tobacco plants coinoculated with two TMV genotypes. A simple model, based on the probability that a leaf in coinoculated plants is infected by just one genotype and on the frequency of each genotype in the source, was used to estimate the effective number of founders for the populations in each leaf. Results from the analysis of three leaves per plant in plants inoculated with different combinations of three TMV genotypes yielded highly consistent estimates. Founder numbers for each leaf were small, in the order of units. This would result in effective population numbers much smaller than the census numbers and indicates that random effects due to genetic drift should be considered for understanding virus evolution within an infected plant.
Figures
Similar articles
-
The multiplicity of infection of a plant virus varies during colonization of its eukaryotic host.J Virol. 2009 Aug;83(15):7487-94. doi: 10.1128/JVI.00636-09. Epub 2009 May 27. J Virol. 2009. PMID: 19474097 Free PMC article.
-
The 2b silencing suppressor of a mild strain of Cucumber mosaic virus alone is sufficient for synergistic interaction with Tobacco mosaic virus and induction of severe leaf malformation in 2b-transgenic tobacco plants.Mol Plant Microbe Interact. 2011 Jun;24(6):685-93. doi: 10.1094/MPMI-12-10-0290. Mol Plant Microbe Interact. 2011. PMID: 21341985
-
Effect of chitosan on tobacco mosaic virus (TMV) accumulation, hydrolase activity, and morphological abnormalities of the viral particles in leaves of N. tabacum L. cv. Samsun.Virol Sin. 2014 Aug;29(4):250-6. doi: 10.1007/s12250-014-3452-8. Epub 2014 Aug 4. Virol Sin. 2014. PMID: 25116808 Free PMC article.
-
Mitochondrial alternative oxidase is involved in both compatible and incompatible host-virus combinations in Nicotiana benthamiana.Plant Sci. 2015 Oct;239:26-35. doi: 10.1016/j.plantsci.2015.07.009. Epub 2015 Jul 19. Plant Sci. 2015. PMID: 26398788
-
Limitations to tobacco mosaic virus infection of turnip.Arch Virol. 1999;144(5):957-71. doi: 10.1007/s007050050558. Arch Virol. 1999. PMID: 10416377
Cited by
-
Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing.Infect Immun. 2013 Jul;81(7):2347-57. doi: 10.1128/IAI.00266-13. Epub 2013 Apr 22. Infect Immun. 2013. PMID: 23608706 Free PMC article.
-
Zucchini yellow mosaic virus (ZYMV, Potyvirus): vertical transmission, seed infection and cryptic infections.Virus Res. 2013 Sep;176(1-2):259-64. doi: 10.1016/j.virusres.2013.06.016. Epub 2013 Jul 8. Virus Res. 2013. PMID: 23845301 Free PMC article.
-
Genetic variability and evolutionary dynamics of viruses of the family Closteroviridae.Front Microbiol. 2013 Jun 26;4:151. doi: 10.3389/fmicb.2013.00151. eCollection 2013. Front Microbiol. 2013. PMID: 23805130 Free PMC article.
-
Challenging the role of microtubules in Tobacco mosaic virus movement by drug treatments is disputable.J Virol. 2006 Jul;80(13):6712-5. doi: 10.1128/JVI.00453-06. J Virol. 2006. PMID: 16775361 Free PMC article.
-
Effects of potyvirus effective population size in inoculated leaves on viral accumulation and the onset of symptoms.J Virol. 2012 Sep;86(18):9737-47. doi: 10.1128/JVI.00909-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740417 Free PMC article.
References
-
- Bruening, G., R. N. Beachy, R. Scalla, and M. Zailtin. 1976. In vitro and in vivo translation of the ribonucleic acids of a cowpea strain of tobacco mosaic virus. Virology 71:498-517. - PubMed
-
- Crow, J. F., and M. Kimura. 1970. An introduction to population genetics theory. Harper & Row, Publishers, Inc., New York, N.Y.
-
- Culver, J. N., and W. O. Dawson. 1989. Point mutations in the coat protein gene of Tobacco mosaic virus induce hypersensitivity in Nicotiana sylvestris. Mol. Plant-Microbe Interact. 2:209-213.
-
- Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

