cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts
- PMID: 12941908
- PMCID: PMC224592
- DOI: 10.1128/jvi.77.18.9979-9986.2003
cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts
Abstract
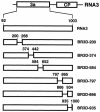
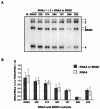
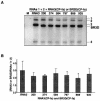
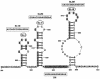
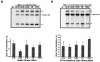
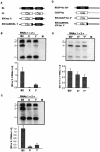
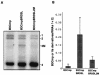
Brome mosaic virus (BMV) is a positive-sense RNA plant virus, the tripartite genomic RNAs of which are separately packaged into virions. RNA3 is copackaged with subgenomic RNA4. In barley protoplasts coinoculated with RNA1 and RNA2, an RNA3 mutant with a 69-nucleotide (nt) deletion in the 3'-proximal region of the 3a open reading frame (ORF) was very poorly packaged compared with other RNA3 mutants and wild-type RNA3, despite their comparable accumulation in the absence of coat protein. Computer analysis of RNA secondary structure predicted two stem-loop (SL) structures (i.e., SL-I and SL-II) in the 69-nt region. Disruption of SL-II, but not of SL-I, significantly reduced RNA3 packaging. A chimeric BMV RNA3 (B3Cmp), with the BMV 3a ORF replacing that of cucumber mosaic virus (CMV), was packaged negligibly, whereas RNA4 was packaged efficiently. Replacement of the 3'-proximal region of the CMV 3a ORF in B3Cmp with the 3'-proximal region of the BMV 3a ORF significantly improved packaging efficiency, and the disruption of SL-II in the substituted BMV 3a ORF region greatly reduced packaging efficiency. These results suggest that the 3'-proximal region of the BMV 3a ORF, especially SL-II predicted between nt 904 and 933, plays an important role in the packaging of BMV RNA3 in vivo. Furthermore, the efficient packaging of RNA4 without RNA3 in B3Cmp-infected cells implies the presence of an element in the 3a ORF of BMV RNA3 that regulates the copackaging of RNA3 and RNA4.
Figures
References
-
- Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. - PubMed
-
- Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. - PubMed
-
- Callaway, A., D. Giesman-Cookmeyer, E. T. Gillock, T. L. Sit, and S. A. Lommel. 2001. The multifunctional capsid protein of plant RNA viruses. Annu. Rev. Phytopathol. 39:419-460. - PubMed
-
- Choi, Y. G., G. L. Grantham, and A. L. N. Rao. 2000. Molecular studies on bromovirus capsid protein. VI. Contributions of the N-terminal arginine-rich motif of BMV capsid protein to virion stability and RNA packaging. Virology 270:377-385. - PubMed
-
- Choi, Y. G., and A. L. N. Rao. 2000. Molecular studies on bromovirus capsid protein. VII. Selective packaging of BMV RNA4 by specific N-terminal arginine residues. Virology 275:207-217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

