Development of pseudorabies virus strains expressing red fluorescent proteins: new tools for multisynaptic labeling applications
- PMID: 12941921
- PMCID: PMC224573
- DOI: 10.1128/jvi.77.18.10106-10112.2003
Development of pseudorabies virus strains expressing red fluorescent proteins: new tools for multisynaptic labeling applications
Abstract
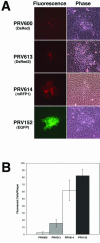
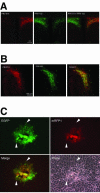
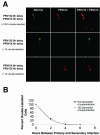
The transsynaptic retrograde transport of the pseudorabies virus Bartha (PRV-Bartha) strain has become an important neuroanatomical tract-tracing technique. Recently, dual viral transneuronal labeling has been introduced by employing recombinant strains of PRV-Bartha engineered to express different reporter proteins. Dual viral transsynaptic tracing has the potential of becoming an extremely powerful method for defining connections of single neurons to multiple neural circuits in the brain. However, the present use of recombinant strains of PRV expressing different reporters that are driven by different promoters, inserted in different regions of the viral genome, and detected by different methods limits the potential of these recombinant virus strains as useful reagents. We previously constructed and characterized PRV152, a PRV-Bartha derivative that expresses the enhanced green fluorescent protein. The development of a strain isogenic to PRV152 and differing only in the fluorescent reporter would have great utility for dual transsynaptic tracing. In this report, we describe the construction, characterization, and application of strain PRV614, a PRV-Bartha derivative expressing a novel monomeric red fluorescent protein, mRFP1. In contrast to viruses expressing DsRed and DsRed2, PRV614 displayed robust fluorescence both in cell culture and in vivo following transsynaptic transport through autonomic circuits afferent to the eye. Transneuronal retrograde dual PRV labeling has the potential to be a powerful addition to the neuroanatomical tools for investigation of neuronal circuits; the use of strain PRV614 in combination with strain PRV152 will eliminate many of the pitfalls associated with the presently used pairs of PRV recombinants.
Figures

References
-
- Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83-87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

