Concerted simulations reveal how peroxidase compound III formation results in cellular oscillations
- PMID: 12944259
- PMCID: PMC1303318
- DOI: 10.1016/S0006-3495(03)74574-3
Concerted simulations reveal how peroxidase compound III formation results in cellular oscillations
Abstract
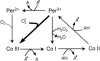
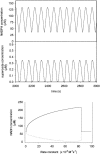
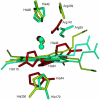
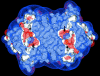
A major problem in mathematical modeling of the dynamics of complex biological systems is the frequent lack of knowledge of kinetic parameters. Here, we apply Brownian dynamics simulations, based on protein three-dimensional structures, to estimate a previously undetermined kinetic parameter, which is then used in biochemical network simulations. The peroxidase-oxidase reaction involves many elementary steps and displays oscillatory dynamics important for immune response. Brownian dynamics simulations were performed for three different peroxidases to estimate the rate constant for one of the elementary steps crucial for oscillations in the peroxidase-oxidase reaction, the association of superoxide with peroxidase. Computed second-order rate constants agree well with available experimental data and permit prediction of rate constants at physiological conditions. The simulations show that electrostatic interactions depress the rate of superoxide association with myeloperoxidase, bringing it into the range necessary for oscillatory behavior in activated neutrophils. Such negative electrostatic steering of enzyme-substrate association presents a novel control mechanism and lies in sharp contrast to the electrostatically-steered fast association of superoxide and Cu/Zn superoxide dismutase, which is also simulated here. The results demonstrate the potential of an integrated and concerted application of structure-based simulations and biochemical network simulations in cellular systems biology.
Figures

References
-
- Amit, A., A. L. Kindzelskii, J. Zanoni, J. N. Jarvis, and H. R. Petty. 1999. Complement deposition on immune complexes reduces the frequencies of metabolic, proteolytic and superoxide oscillations of migrating neutrophils. Cell. Immunol. 194:47–53. - PubMed
-
- Berglund, G. I., G. H. Carlsson, A. T. Smith, H. Szöke, A. Henriksen, and J. Hajdu. 2002. The catalytic pathway of horseradish peroxidase at high resolution. Nature. 417:463–467. - PubMed
-
- Demchuk, E., and R. C. Wade. 1996. Improving the continuum dielectric approach to calculating pKas of ionizable groups in proteins. J. Phys. Chem. 100:17373–17387.
-
- Elcock, A. H., R. R. Gabdoulline, R. C. Wade, and J. A. McCammon. 1999. Computer simulation of protein-protein association kinetics: acetylcholinesterase-fasciculin. J. Mol. Biol. 291:149–162. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

