Formation pathways of a guanine-quadruplex DNA revealed by molecular dynamics and thermodynamic analysis of the substates
- PMID: 12944293
- PMCID: PMC1303352
- DOI: 10.1016/S0006-3495(03)74608-6
Formation pathways of a guanine-quadruplex DNA revealed by molecular dynamics and thermodynamic analysis of the substates
Abstract
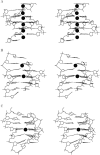
The formation of a cation-stabilized guanine quadruplex (G-DNA) stem is an exceptionally slow process involving complex kinetics that has not yet been characterized at atomic resolution. Here, we investigate the formation of a parallel stranded G-DNA stem consisting of four strands of d(GGGG) using molecular dynamics simulations with explicit inclusion of counterions and solvent. Due to the limitations imposed by the nanosecond timescale of the simulations, rather than watching for the spontaneous formation of G-DNA, our approach probes the stability of possible supramolecular intermediates (including two-, three-, and four-stranded assemblies with out-of-register base pairing between guanines) on the formation pathway. The simulations suggest that "cross-like" two-stranded assemblies may serve as nucleation centers in the initial formation of parallel stranded G-DNA quadruplexes, proceeding through a series of rearrangements involving trapping of cations, association of additional strands, and progressive slippage of strands toward the full stem. To supplement the analysis, approximate free energies of the models are obtained with explicit consideration of the integral cations. The approach applied here serves as a prototype for qualitatively investigating other G-DNA molecules using molecular dynamics simulation and free-energy analysis.
Figures



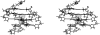
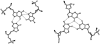



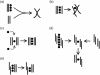


References
-
- Andricioaei, I., and M. Karplus. 2001. On the calculation of the entropy from covariance matrices on the atomic fluctuations. J. Chem. Phys. 115:6289–6292.
-
- Aqvist, J. 1990. Ion-water interaction potentials derived from free energy perturbation simulations. J. Phys. Chem. 94:8021–8024.
-
- Arnott, S., and D. W. Hukins. 1972. Optimised parameters for A-DNA and B-DNA. Biochem. Biophys. Res. Commun. 47:1504–1509. - PubMed
-
- Arthanari, H., and P. H. Bolton. 1999. Porphyrins can catalyze the interconversion of DNA quadruplex structural types. Anticancer Drug Des. 14:317–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

