Identification of binding mechanisms in single molecule-DNA complexes
- PMID: 12944309
- PMCID: PMC1303368
- DOI: 10.1016/S0006-3495(03)74624-4
Identification of binding mechanisms in single molecule-DNA complexes
Abstract
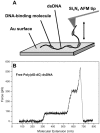
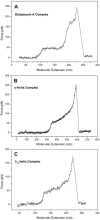
Changes in the elastic properties of single deoxyribonucleic acid (DNA) molecules in the presence of different DNA-binding agents are identified using atomic force microscope single molecule force spectroscopy. We investigated the binding of poly(dG-dC) dsDNA with the minor groove binder distamycin A, two supposed major groove binders, an alpha-helical and a 3(10)-helical peptide, the intercalants daunomycin, ethidium bromide and YO, and the bis-intercalant YOYO. Characteristic mechanical fingerprints in the overstretching behavior of the studied single DNA-ligand complexes were observed allowing the distinction between different binding modes. Docking of ligands to the minor or major groove of DNA has the effect that the intramolecular B-S transition remains visible as a distinct plateau in the force-extension trace. By contrast, intercalation of small molecules into the double helix is characterized by the vanishing of the B-S plateau. These findings lead to the conclusion that atomic force microscope force spectroscopy can be regarded as a single molecule biosensor and is a potent tool for the characterization of binding motives of small ligands to DNA.
Figures

Similar articles
-
Molecular mechanisms and kinetics between DNA and DNA binding ligands.Biophys J. 2005 Jan;88(1):404-11. doi: 10.1529/biophysj.103.036293. Epub 2004 Oct 29. Biophys J. 2005. PMID: 15516529 Free PMC article.
-
DNA interactions of cisplatin tethered to the DNA minor groove binder distamycin.Eur J Biochem. 1999 Dec;266(2):392-402. doi: 10.1046/j.1432-1327.1999.00866.x. Eur J Biochem. 1999. PMID: 10561579
-
Discriminating small molecule DNA binding modes by single molecule force spectroscopy.FEBS Lett. 2002 Jan 16;510(3):154-8. doi: 10.1016/s0014-5793(01)03257-4. FEBS Lett. 2002. PMID: 11801245
-
Drug-DNA sequence-dependent interactions analysed by electric linear dichroism.J Mol Recognit. 1992 Dec;5(4):155-71. doi: 10.1002/jmr.300050406. J Mol Recognit. 1992. PMID: 1339484 Review.
-
Single molecule force spectroscopy on ligand-DNA complexes: from molecular binding mechanisms to biosensor applications.J Biotechnol. 2004 Aug 26;112(1-2):5-12. doi: 10.1016/j.jbiotec.2004.04.029. J Biotechnol. 2004. PMID: 15288936 Review.
Cited by
-
Mechanisms of small molecule-DNA interactions probed by single-molecule force spectroscopy.Nucleic Acids Res. 2016 May 19;44(9):3971-88. doi: 10.1093/nar/gkw237. Epub 2016 Apr 16. Nucleic Acids Res. 2016. PMID: 27085806 Free PMC article.
-
Interaction of synthetic antimicrobial peptides of the Hylin a1 family with models of eukaryotic structures: Zwitterionic membranes and DNA.Biochem Biophys Rep. 2020 Nov 3;24:100827. doi: 10.1016/j.bbrep.2020.100827. eCollection 2020 Dec. Biochem Biophys Rep. 2020. PMID: 33195825 Free PMC article.
-
ZYH005, a novel DNA intercalator, overcomes all-trans retinoic acid resistance in acute promyelocytic leukemia.Nucleic Acids Res. 2018 Apr 20;46(7):3284-3297. doi: 10.1093/nar/gky202. Nucleic Acids Res. 2018. PMID: 29554366 Free PMC article.
-
Molecular Dynamics and Multi-Spectroscopic of the Interaction Behavior between Bladder Cancer Cells and Calf Thymus DNA with Rebeccamycin: Apoptosis through the Down Regulation of PI3K/AKT Signaling Pathway.J Fluoresc. 2023 Jul;33(4):1537-1557. doi: 10.1007/s10895-023-03169-4. Epub 2023 Feb 14. J Fluoresc. 2023. PMID: 36787038
-
Exploring the interaction of ruthenium(II) polypyridyl complexes with DNA using single-molecule techniques.Langmuir. 2006 May 9;22(10):4699-709. doi: 10.1021/la053242r. Langmuir. 2006. PMID: 16649785 Free PMC article.
References
-
- Anselmetti, D., J. Fritz, B. Smith, and X. Fernandez-Busquets. 2000. Single molecule DNA biophysics with atomic force microscopy. Single Molecules. 1:17–23.
-
- Aubel-Sadron, G., and D. Londos-Gagliardi. 1984. Daunorubicin and doxorubicin, anthracycline antibiotics, a physicochemical and biological review. Biochimie. 66:333–352. - PubMed
-
- Baguley, B. C. 1982. Nonintercalative DNA-binding antitumour compounds. Mol. Cell. Biochem. 43:167–181. - PubMed
-
- Bailly, C., and J. B. Chaires. 1998. Sequence-specific DNA minor groove binders. Design and synthesis of netropsin and distamycin analogues. Bioconjug. Chem. 9:513–538. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

