Rad52-independent accumulation of joint circular minichromosomes during S phase in Saccharomyces cerevisiae
- PMID: 12944465
- PMCID: PMC193689
- DOI: 10.1128/MCB.23.18.6363-6372.2003
Rad52-independent accumulation of joint circular minichromosomes during S phase in Saccharomyces cerevisiae
Abstract
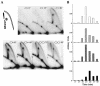
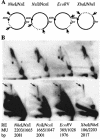
We investigated the formation of X-shaped molecules consisting of joint circular minichromosomes (joint molecules) in Saccharomyces cerevisiae by two-dimensional neutral/neutral gel electrophoresis of psoralen-cross-linked DNA. The appearance of joint molecules was found to be replication dependent. The joint molecules had physical properties reminiscent of Holliday junctions or hemicatenanes, as monitored by strand displacement, branch migration, and nuclease digestion. Physical linkage of the joint molecules was detected along the entire length of the minichromosome and most likely involved newly replicated sister chromatids. Surprisingly, the formation of joint molecules was found to be independent of Rad52p as well as of other factors associated with a function in homologous recombination or in the resolution of stalled replication intermediates. These findings thus imply the existence of a nonrecombinational pathway(s) for the formation of joint molecules during the process of DNA replication or minichromosome segregation.
Figures




Similar articles
-
Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms.Mol Cell. 2003 Dec;12(6):1499-510. doi: 10.1016/s1097-2765(03)00473-8. Mol Cell. 2003. PMID: 14690603
-
Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint.EMBO J. 2009 Apr 22;28(8):1131-41. doi: 10.1038/emboj.2009.75. Epub 2009 Mar 26. EMBO J. 2009. PMID: 19322196 Free PMC article.
-
A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52.Mol Cell Biol. 1995 Mar;15(3):1620-31. doi: 10.1128/MCB.15.3.1620. Mol Cell Biol. 1995. PMID: 7862153 Free PMC article.
-
Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae.Microbiol Mol Biol Rev. 1999 Jun;63(2):349-404. doi: 10.1128/MMBR.63.2.349-404.1999. Microbiol Mol Biol Rev. 1999. PMID: 10357855 Free PMC article. Review.
-
The S-phase checkpoint and its regulation in Saccharomyces cerevisiae.Mutat Res. 2003 Nov 27;532(1-2):41-58. doi: 10.1016/j.mrfmmm.2003.08.009. Mutat Res. 2003. PMID: 14643428 Review.
Cited by
-
Impaired manganese metabolism causes mitotic misregulation.J Biol Chem. 2012 May 25;287(22):18717-29. doi: 10.1074/jbc.M112.358309. Epub 2012 Apr 4. J Biol Chem. 2012. PMID: 22493290 Free PMC article.
-
An hpr1 point mutation that impairs transcription and mRNP biogenesis without increasing recombination.Mol Cell Biol. 2006 Oct;26(20):7451-65. doi: 10.1128/MCB.00684-06. Epub 2006 Aug 14. Mol Cell Biol. 2006. PMID: 16908536 Free PMC article.
-
Partial depletion of histone H4 increases homologous recombination-mediated genetic instability.Mol Cell Biol. 2005 Feb;25(4):1526-36. doi: 10.1128/MCB.25.4.1526-1536.2005. Mol Cell Biol. 2005. PMID: 15684401 Free PMC article.
-
Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase.Genes Dev. 2005 Feb 1;19(3):339-50. doi: 10.1101/gad.322605. Genes Dev. 2005. PMID: 15687257 Free PMC article.
-
Construction of DNA hemicatenanes from two small circular DNA molecules.PLoS One. 2015 Mar 23;10(3):e0119368. doi: 10.1371/journal.pone.0119368. eCollection 2015. PLoS One. 2015. PMID: 25799010 Free PMC article.
References
-
- Bai, Y., and L. S. Symington. 1996. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025-2037. - PubMed
-
- Bates, A. D., and A. Maxwell. 1993. DNA topology. Oxford University Press, Oxford, UK.
-
- Bell, L., and B. Byers. 1983. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Anal. Biochem. 130:527-535. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
