PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression
- PMID: 12944487
- PMCID: PMC193712
- DOI: 10.1128/MCB.23.18.6618-6630.2003
PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression
Abstract
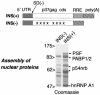
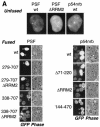
Human immunodeficiency virus type 1 (HIV) gag/pol and env mRNAs contain cis-acting regulatory elements (INS) that impair stability, nucleocytoplasmic transport, and translation by unknown mechanisms. This downregulation can be counteracted by the viral Rev protein, resulting in efficient export and expression of these mRNAs. Here, we show that the INS region in HIV-1 gag mRNA is a high-affinity ligand of p54nrb/PSF, a heterodimeric transcription/splicing factor. Both subunits bound INS RNA in vitro with similar affinity and specificity. Using an INS-containing subgenomic gag mRNA, we show that it specifically associated with p54nrb in vivo and that PSF inhibited its expression, acting via INS. Studying the authentic HIV-1 mRNAs produced from an infectious molecular clone, we found that PSF affected specifically the INS-containing, Rev-dependent transcripts encoding Gag-Pol and Env. Both subunits contained nuclear export and nuclear retention signals, whereas p54nrb was continuously exported from the nucleus and associated with INS-containing mRNA in the cytoplasm, suggesting its additional role at late steps of mRNA metabolism. Thus, p54nrb and PSF have properties of key factors mediating INS function and likely define a novel mRNA regulatory pathway that is hijacked by HIV-1.
Figures

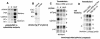





References
-
- Afonina, E., M. Neumann, and G. N. Pavlakis. 1997. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 272:2307-2311. - PubMed
-
- Afonina, E., R. Stauber, and G. N. Pavlakis. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273:13015-13021. - PubMed
-
- Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. - PubMed
-
- Arrigo, S. J., and I. S. Y. Chen. 1991. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 5:808-819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
