Novel insights into the regulation of the timeless protein
- PMID: 12944510
- PMCID: PMC6740606
- DOI: 10.1523/JNEUROSCI.23-21-07810.2003
Novel insights into the regulation of the timeless protein
Abstract
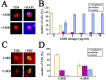
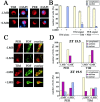
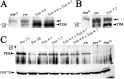
In the Drosophila circadian clock, period (per) and its partner, timeless (tim), play a central role in the negative limb of an autoregulatory feedback loop. Unlike per, the dosage of which affects the frequency (tau) of the circadian cycle, we found that increasing copies of the tim gene has no effect on clock period length. The use of the tim promoter to express per results in a shortening of circadian period, also indicating that the regulation of tim is different from that of per. Drosophila TIM is similar to the mammalian circadian protein mPER2 in that it shuttles independently between the nucleus and cytoplasm both in vivo and in vitro. Contrary to the current model that PER and TIM heterodimerization is a prerequisite for their nuclear entry, PER is not required to transport TIM into nuclei, although it influences TIM localization and vice versa. Blocking nuclear export led to increased nuclear expression of TIM in S2 cells and in wild-type and per01 larvae, suggesting that PER may be required for nuclear retention of TIM. Unlike PER, nuclear TIM alone has no ability to repress transcription. We propose that TIM drives cycles of PER expression by regulating its stability, and in turn, PER retains TIM in the nucleus, either for the regulation of its own stability or for a novel nuclear role of TIM.
Figures



References
-
- Baylies MK, Bargielli TA, Jackson FR, Young MW ( 1987) Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature 326: 390-392. - PubMed
-
- Chen Y, Hunter-Ensor M, Schotland P, Sehgal A ( 1998) Alterations of per RNA in noncoding regions affect periodicity of circadian behavioral rhythms. J Biol Rhythms 13: 364-379. - PubMed
-
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young M ( 2001) Circadian regulation of gene expression systems in the Drosophila head. Neuron 32: 657-671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
