Overproduction of inactive variants of the murein synthase PBP1B causes lysis in Escherichia coli
- PMID: 12949085
- PMCID: PMC193747
- DOI: 10.1128/JB.185.18.5342-5348.2003
Overproduction of inactive variants of the murein synthase PBP1B causes lysis in Escherichia coli
Abstract
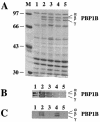
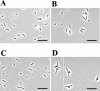
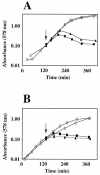
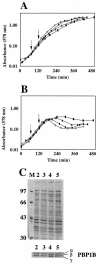
Penicillin-binding protein 1B (PBP1B) of Escherichia coli is a bifunctional murein synthase containing both a transpeptidase domain and a transglycosylase domain. The protein is present in three forms (alpha, beta, and gamma) which differ in the length of their N-terminal cytoplasmic region. Expression plasmids allowing the production of native PBP1B or of PBP1B variants with an inactive transpeptidase or transglycosylase domain or both were constructed. The inactive domains contained a single amino acid exchange in an essential active-site residue. Overproduction of the inactive PBP1B variants, but not of the active proteins, caused lysis of wild-type cells. The cells became tolerant to lysis by inactive PBP1B at a pH of 5.0, which is similar to the known tolerance for penicillin-induced lysis under acid pH conditions. Lysis was also reduced in mutant strains lacking several murein hydrolases. In particular, a strain devoid of activity of all known lytic transglycosylases was virtually tolerant, indicating that mainly the lytic transglycosylases are responsible for the observed lysis effect. A possible structural interaction between PBP1B and murein hydrolases in vivo by the formation of a multienzyme complex is discussed.
Figures


References
-
- Bishop, R. E., and J. H. Weiner. 1993. Complementation of growth defect in an ampC deletion mutant of Escherichia coli. FEMS Microbiol. Lett. 114:349-354. - PubMed
-
- Casabadan, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

