Mycobacterium tuberculosis FurA autoregulates its own expression
- PMID: 12949087
- PMCID: PMC193761
- DOI: 10.1128/JB.185.18.5357-5362.2003
Mycobacterium tuberculosis FurA autoregulates its own expression
Abstract
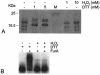
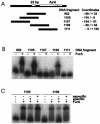
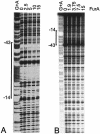
The furA-katG region of Mycobacterium tuberculosis, encoding a Fur-like protein and the catalase-peroxidase, is highly conserved among mycobacteria. Both genes are induced upon oxidative stress. In this work we analyzed the M. tuberculosis furA promoter region. DNA fragments were cloned upstream of the luciferase reporter gene, and promoter activity in Mycobacterium smegmatis was measured in both the presence and absence of oxidative stress. The shortest fragment containing an inducible promoter extends 45 bp upstream of furA. In this region, -35 and -10 promoter consensus sequences can be identified, as well as a 23-bp AT-rich sequence that is conserved in the nonpathogenic but closely related M. smegmatis. M. tuberculosis FurA was purified and found to bind upstream of furA by gel shift analysis. A ca. 30-bp DNA sequence, centered on the AT-rich region, was essential for FurA binding and protected by FurA in footprinting analysis. Peroxide treatment of FurA abolished DNA binding. Three different AT-rich sequences mutagenized by site-directed mutagenesis were constructed. In each mutant, both M. tuberculosis FurA binding in vitro and pfurA regulation upon oxidative-stress in M. smegmatis were abolished. Thus, pfurA is an oxidative stress-responsive promoter controlled by the FurA protein.
Figures



References
-
- Althaus, E. W., C. E. Outten, K. E. Olson, H. Cao, and T. V. Halloran. 1999. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry 38:6559-6569. - PubMed
-
- Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. - PubMed
-
- Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. - PubMed
-
- Bullock, W., J. Fernandez, and J. Short. 1987. XLI-Blue: a high efficiency plasmid transforming recA E. coli strain with β-galactosidase selection. BioTechniques 5:376-379.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

