Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar enteritidis
- PMID: 12949092
- PMCID: PMC193744
- DOI: 10.1128/JB.185.18.5398-5407.2003
Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar enteritidis
Abstract
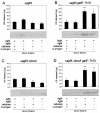
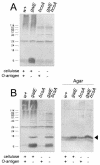
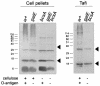
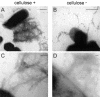
Lipopolysaccharide (LPS) O polysaccharide was identified as the principle factor impeding intercellular formation of intact thin aggregative fimbriae (Tafi) in Salmonella enterica serovar Enteritidis. The extracellular nucleation-precipitation assembly pathway for these organelles was investigated by quantifying fimbrial formation between deltaagfA (AgfA recipient) and deltaagfB (AgfA donor) cells harboring mutations in LPS (galE::Tn10) and/or cellulose (deltabcsA) synthesis. Intercellular complementation could be detected between deltaagfA and deltaagfB strains only when both possessed the galE mutation. LPS O polysaccharide appears to be an impenetrable barrier to AgfA assembly between cells but not within individual cells. The presence of cellulose did not restrict Tafi formation between cells. Transmission electron microscopy of w+ S. enterica serovar Enteritidis 3b cells revealed diffuse Tafi networks without discernible fine structure. In the absence of cellulose, however, individual Tafi fibers were clearly visible, appeared to be occasionally branched, and showed the generally distinctive appearance described for Escherichia coli K-12 curli. A third extracellular matrix component closely associated with cellulose and Tafi was detected on Western blots by using immune serum raised to whole, purified Tafi aggregates. Cellulose was required to tightly link this material to cells. Antigenically similar material was also detected in S. enterica serovar Typhimurium and one diarrheagenic E. coli isolate. Preliminary analysis indicated that this material represented an anionic, extracellular polysaccharide that was distinct from colanic acid. Therefore, Tafi in their native state appear to exist as a complex with cellulose and at least one other component.
Figures

References
-
- Aguilar, A., S. Merino, M. M. Nogueras, M. Regue, and J. M. Tomas. 1999. Two genes from the capsule of Aeromonas hydrophila (serogroup O:34) confer serum resistance to Escherichia coli K12 strains. Res. Microbiol. 150:395-402. - PubMed
-
- Arnqvist, A., A. Olsen, J. Pfeifer, D. G. Russell, and S. Normark. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol. Microbiol. 6:2443-2452. - PubMed
-
- Austin, J. W., G. Sanders, W. W. Kay, and S. K. Collinson. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162:295-301. - PubMed
-
- Baumler, A. J., A. J. Gilde, R. M. Tsolis, A. W. van der Velden, B. M. Ahmer, and F. Heffron. 1997. Contribution of horizontal gene transfer and deletion events to development of distinctive patterns of fimbrial operons during evolution of Salmonella serotypes. J. Bacteriol. 179:317-322. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

