Natural transformation of Campylobacter jejuni requires components of a type II secretion system
- PMID: 12949093
- PMCID: PMC193740
- DOI: 10.1128/JB.185.18.5408-5418.2003
Natural transformation of Campylobacter jejuni requires components of a type II secretion system
Erratum in
- J Bacteriol. 2003 Nov;185(21):6493
Abstract
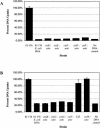
The human pathogen Campylobacter jejuni is one of more than 40 naturally competent bacterial species able to import macromolecular DNA from the environment and incorporate it into their genomes. However, in C. jejuni little is known about the genes involved in this process. We used random transposon mutagenesis to identify genes that are required for the transformation of this organism. We isolated mutants with insertions in 11 different genes; most of the mutants are affected in the DNA uptake stage of transformation, whereas two mutants are affected in steps subsequent to DNA uptake, such as recombination into the chromosome or in DNA transport across the inner membrane. Several of these genes encode proteins homologous to those involved in type II secretion systems, biogenesis of type IV pili, and competence for natural transformation in gram-positive and gram-negative species. Other genes identified in our screen encode proteins unique to C. jejuni or are homologous to proteins that have not been shown to play a role in the transformation in other bacteria.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

