Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus
- PMID: 12949095
- PMCID: PMC193756
- DOI: 10.1128/JB.185.18.5431-5441.2003
Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus
Abstract
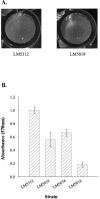
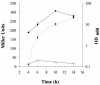
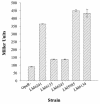
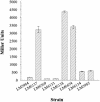
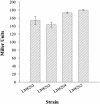
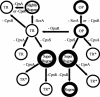
Vibrio parahaemolyticus, a biofouling marine bacterium and human pathogen, undergoes phase variation displaying translucent (TR) and opaque (OP) colony morphologies. Prior studies demonstrated that OP colonies produce more capsular polysaccharide (CPS) than TR colonies and that opacity is controlled by the Vibrio harveyi LuxR-type transcriptional activator OpaR. CPS has also been shown to be regulated by the scrABC signaling pathway, which involves a GGDEF-EAL motif-containing sensory protein. The present study identifies cps genes and examines their regulation. Transposon insertions in the cps locus, which contains 11 genes, abolished opacity. Such mutants failed to produce CPS and were defective in pellicle formation in microtiter wells and in a biofilm attachment assay. Reporter fusions to cpsA, the first gene in the locus, showed approximately 10-fold-enhanced transcription in the OP (opaR+) strain compared to a TR (deltaopaR) strain. Two additional transcriptional regulators were discovered. One potential activator, CpsR, participates in the scrABC GGDEF-EAL-signaling pathway; CpsR was required for the increased cps expression observed in scrA deltaopaR strains. CpsR, which contains a conserved module found in members of the AAA+ superfamily of ATP-interacting proteins, is homologous to Vibrio cholerae VpsR; however, unlike VpsR, CpsR was not essential for cps expression. CpsS, the second newly identified regulator, contains a CsgD-type DNA-binding domain and appears to act as a repressor. Mutants with cpsS defects have greatly elevated cps transcription; their high level of cpsA expression was CpsR dependent in TR strains and primarily OpaR dependent in OP strains. Thus, a network of positive and negative regulators modulates CPS production in V. parahaemolyticus.
Figures
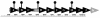


References
-
- Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

