Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations
- PMID: 12949110
- PMCID: PMC193770
- DOI: 10.1128/JB.185.18.5573-5584.2003
Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations
Abstract
Thirteen different serotypes of Listeria monocytogenes can be distinguished on the basis of variation in somatic and flagellar antigens. Although the known virulence genes are present in all serotypes, greater than 90% of human cases of listeriosis are caused by serotypes 1/2a, 1/2b, and 4b and nearly all outbreaks of food-borne listeriosis have been caused by serotype 4b strains. Phylogenetic analysis of these three common clinical serotypes places them into two different lineages, with serotypes 1/2b and 4b belonging to lineage I and 1/2a belonging to lineage II. To begin examining evolution of the genome in these serotypes, DNA microarray analysis was used to identify lineage-specific and serotype-specific differences in genome content. A set of 44 strains representing serotypes 1/2a, 1/2b, and 4b was probed with a shotgun DNA microarray constructed from the serotype 1/2a strain 10403s. Clones spanning 47 different genes in 16 different contiguous segments relative to the lineage II 1/2a genome were found to be absent in all lineage I strains tested (serotype 4b and 1/2b) and an additional nine were altered exclusively in 4b strains. Southern hybridization confirmed that conserved alterations were, in all but two loci, due to absence of the segments from the genome. Genes within these contiguous segments comprise five functional categories, including genes involved in synthesis of cell surface molecules and regulation of virulence gene expression. Phylogenetic reconstruction and examination of compositional bias in the regions of difference are consistent with a model in which the ancestor of the two lineages had the 1/2 somatic serotype and the regions absent in the lineage I genome arose by loss of ancestral sequences.
Figures
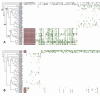



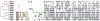

References
-
- Aarts, H. J., L. E. Hakemulder, and A. M. Van Hoef. 1999. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int. J. Food Microbiol. 49:95-102. - PubMed
-
- Bibb, W. F., B. Schwartz, B. G. Gellin, B. D. Plikaytis, and R. E. Weaver. 1989. Analysis of Listeria monocytogenes by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Int. J. Food Microbiol. 8:233-239. - PubMed
-
- Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

