Severe acute respiratory syndrome--a new coronavirus from the Chinese dragon's lair
- PMID: 12950672
- PMCID: PMC7169508
- DOI: 10.1046/j.1365-3083.2003.01302.x
Severe acute respiratory syndrome--a new coronavirus from the Chinese dragon's lair
Abstract
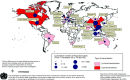
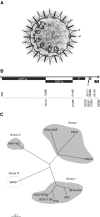
The recent identification of a novel clinical entity, the severe acute respiratory syndrome (SARS), the rapid subsequent spread and case fatality rates of 14-15% have prompted a massive international collaborative investigation facilitated by a network of laboratories established by the World Health Organization (WHO). As SARS has the potential of becoming the first pandemic of the new millennium, a global warning by the WHO was issued on 12 March 2003. The disease, which is believed to have its origin in the Chinese Guangdong province, spread from Hong Kong via international airports to its current worldwide distribution. The concerted efforts of a globally united scientific community have led to the independent isolation and identification of a novel coronavirus from SARS patients by several groups. The extraordinarily rapid isolation of a causative agent of this newly emerged infectious disease constitutes an unprecedented scientific achievement. The main scope of the article is to provide the clinician with an overview of the natural history, epidemiology and clinical characteristics of SARS. On the basis of the recently published viral genome and structural features common to the members of the coronavirus family, a model for host cell-virus interaction and possible targets for antiviral drugs are presented. The epidemiological consequences of introducing a novel pathogen in a previously unexposed population and the origin and evolution of a new and more pathogenic strain of coronavirus are discussed.
Figures
Similar articles
-
Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome.Lancet. 2004 Jan 10;363(9403):99-104. doi: 10.1016/s0140-6736(03)15259-2. Lancet. 2004. PMID: 14726162 Free PMC article.
-
Severe acute respiratory syndrome (SARS): breath-taking progress.J Med Microbiol. 2003 Aug;52(Pt 8):609-613. doi: 10.1099/jmm.0.05321-0. J Med Microbiol. 2003. PMID: 12867552 Review.
-
Epidemiology of severe acute respiratory syndrome (SARS): adults and children.Paediatr Respir Rev. 2004 Dec;5(4):270-4. doi: 10.1016/j.prrv.2004.07.011. Paediatr Respir Rev. 2004. PMID: 15531250 Free PMC article. Review.
-
A multicentre collaboration to investigate the cause of severe acute respiratory syndrome.Lancet. 2003 May 17;361(9370):1730-3. doi: 10.1016/s0140-6736(03)13376-4. Lancet. 2003. PMID: 12767752 Free PMC article.
-
Overview: The history and pediatric perspectives of severe acute respiratory syndromes: Novel or just like SARS.Pediatr Pulmonol. 2020 Jul;55(7):1584-1591. doi: 10.1002/ppul.24810. Epub 2020 Jun 1. Pediatr Pulmonol. 2020. PMID: 32483934 Free PMC article. Review.
Cited by
-
SARS vaccines: where are we?Expert Rev Vaccines. 2009 Jul;8(7):887-98. doi: 10.1586/erv.09.43. Expert Rev Vaccines. 2009. PMID: 19538115 Free PMC article. Review.
-
Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: target, mechanism of action, and therapeutic potential.Rev Med Virol. 2012 Jan;22(1):2-17. doi: 10.1002/rmv.706. Epub 2011 Sep 8. Rev Med Virol. 2012. PMID: 21905149 Free PMC article. Review.
-
Molecular advances in severe acute respiratory syndrome-associated coronavirus (SARS-CoV).Genomics Proteomics Bioinformatics. 2003 Nov;1(4):247-62. doi: 10.1016/s1672-0229(03)01031-3. Genomics Proteomics Bioinformatics. 2003. PMID: 15629054 Free PMC article. Review.
-
Human monoclonal antibodies to SARS-coronavirus inhibit infection by different mechanisms.Virology. 2009 Nov 10;394(1):39-46. doi: 10.1016/j.virol.2009.07.028. Epub 2009 Sep 12. Virology. 2009. PMID: 19748648 Free PMC article.
References
-
- WHO. Situation updates – severe acute respiratory syndrome (SARS) update 52 (available at: http://www.who.int).
-
- Lee N, Hui D, Wu A et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003; 348: 1986–94. - PubMed
-
- Booth CM, Matukas LM, Tomlinson GA et al. Clinical features and short‐term outcomes of 144 patients with SARS in the Greater Toronto Area. JAMA 2003;289: 2801–9. - PubMed
-
- Peiris JSM, Chu CM, Cheng, VCC et al. Prospective study of the clinical progression and viral load of SARS associated coronavirus pneumonia in a community outbreak (accepted in Lancet and available at: http://www.who.int). - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

